Alzheimer’s disease (AD) is an irreversibly progressive neurodegenerative disease afflicting the elderly, accompanied by devastating cognitive and memory impairment caused by characteristic neuronal and synaptic loss and cortical and hippocampal atrophy. It is hallmarked by the accumulation of extracellular amyloid plaques and intracellular neurofibrillary tangles. The underlying mechanisms contributing to the development of the disease remain elusive and controversial. Despite the advancement in understanding the mechanism of pathogenesis, clinical trials have been unsuccessful and provided no relief from disease progression, only slowing the progression. Recent FDA-approved anti-amyloid therapy aducanumab highlights that it is effective for patients with very mild, biomarker-proven AD. Therefore, there is an urgent need to develop a more accessible biomarker screening test using less invasive and cost-effective body fluid biomarkers. These diagnostics will serve as the first line of effective AD therapies before extensive pathophysiological brain devastation occurs.
- Alzheimer’s disease
- nasal discharge fluid
- body fluid
- biomarker
- diagnosis
- non-invasive
- peripheral
1. Conventional AD Body Fluid Biomarkers
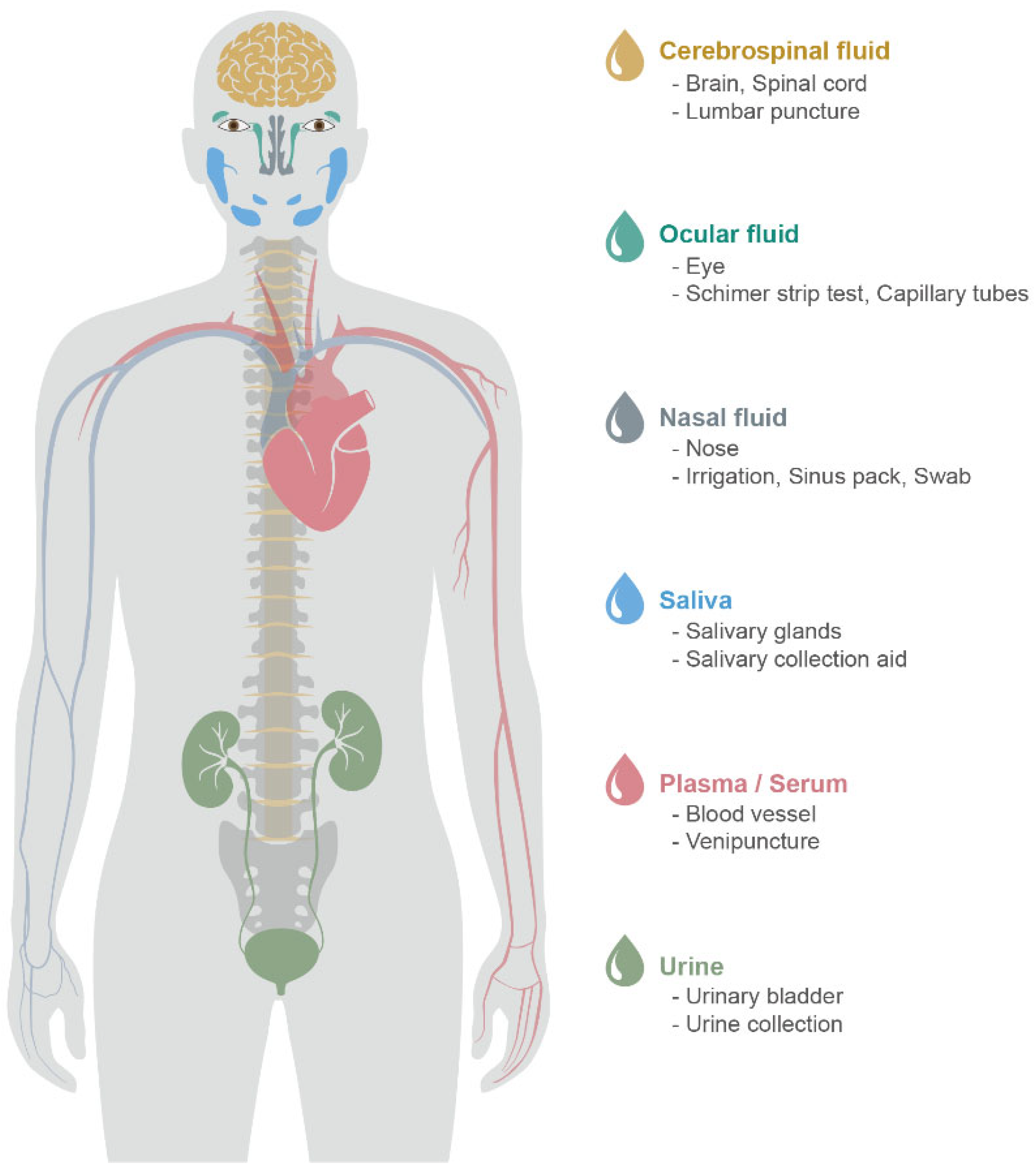
1.1. CSF
1.2. Blood (Plasma and Serum)
2. Novel Peripheral Body Fluid Biomarkers
2.1. Nasal Discharge
2.2. Tears
2.3. Saliva
2.4. Urine
This entry is adapted from the peer-reviewed paper 10.3390/pharmaceutics14081532
References
- Baker, M. In biomarkers we trust? Nat. Biotechnol. 2005, 23, 297–304.
- Ray, P.; Le Manach, Y.; Riou, B.; Houle, T.T. Statistical evaluation of a biomarker. Anesthesiology 2010, 112, 1023–1040.
- Jack, C.R., Jr.; Bennett, D.A.; Blennow, K.; Carrillo, M.C.; Dunn, B.; Haeberlein, S.B.; Holtzman, D.M.; Jagust, W.; Jessen, F.; Karlawish, J.; et al. NIA-AA Research Framework: Toward a biological definition of Alzheimer’s disease. Alzheimers Dement. 2018, 14, 535–562.
- Blennow, K.; Hampel, H.; Weiner, M.; Zetterberg, H. Cerebrospinal fluid and plasma biomarkers in Alzheimer disease. Nat. Rev. Neurol. 2010, 6, 131–144.
- Sakka, L.; Coll, G.; Chazal, J. Anatomy and physiology of cerebrospinal fluid. Eur. Ann. Otorhinolaryngol. Head Neck Dis. 2011, 128, 309–316.
- van Gool, A.J.; Hendrickson, R.C. The proteomic toolbox for studying cerebrospinal fluid. Expert Rev. Proteom. 2012, 9, 165–179.
- Lee, J.C.; Kim, S.J.; Hong, S.; Kim, Y. Diagnosis of Alzheimer’s disease utilizing amyloid and tau as fluid biomarkers. Exp. Mol. Med. 2019, 51, 1–10.
- Rosenberg, G.A. Chapter 4—Cerebrospinal Fluid: Formation, Absorption, Markers, and Relationship to Blood–Brain Barrier. In Primer on Cerebrovascular Diseases, 2nd ed.; Caplan, L.R., Biller, J., Leary, M.C., Lo, E.H., Thomas, A.J., Yenari, M., Zhang, J.H., Eds.; Academic Press: San Diego, CA, USA, 2017; pp. 25–31.
- Teunissen, C.E.; Petzold, A.; Bennett, J.L.; Berven, F.S.; Brundin, L.; Comabella, M.; Franciotta, D.; Frederiksen, J.L.; Fleming, J.O.; Furlan, R.; et al. A consensus protocol for the standardization of cerebrospinal fluid collection and biobanking. Neurology 2009, 73, 1914–1922.
- Cummings, J.L.; Dubois, B.; Molinuevo, J.L.; Scheltens, P. International Work Group criteria for the diagnosis of Alzheimer disease. Med. Clin. N. Am. 2013, 97, 363–368.
- Dubois, B.; Feldman, H.H.; Jacova, C.; Dekosky, S.T.; Barberger-Gateau, P.; Cummings, J.; Delacourte, A.; Galasko, D.; Gauthier, S.; Jicha, G.; et al. Research criteria for the diagnosis of Alzheimer’s disease: Revising the NINCDS-ADRDA criteria. Lancet Neurol. 2007, 6, 734–746.
- Dubois, B.; Feldman, H.H.; Jacova, C.; Hampel, H.; Molinuevo, J.L.; Blennow, K.; DeKosky, S.T.; Gauthier, S.; Selkoe, D.; Bateman, R.; et al. Advancing research diagnostic criteria for Alzheimer’s disease: The IWG-2 criteria. Lancet Neurol. 2014, 13, 614–629.
- Oddo, S.; Caccamo, A.; Kitazawa, M.; Tseng, B.P.; LaFerla, F.M. Amyloid deposition precedes tangle formation in a triple transgenic model of Alzheimer’s disease. Neurobiol. Aging 2003, 24, 1063–1070.
- Braak, H.; Zetterberg, H.; Del Tredici, K.; Blennow, K. Intraneuronal tau aggregation precedes diffuse plaque deposition, but amyloid-β changes occur before increases of tau in cerebrospinal fluid. Acta Neuropathol. 2013, 126, 631–641.
- Zetterberg, H.; Wahlund, L.O.; Blennow, K. Cerebrospinal fluid markers for prediction of Alzheimer’s disease. Neurosci. Lett. 2003, 352, 67–69.
- Krebs, H.A. Chemical composition of blood plasma and serum. Annu. Rev. Biochem. 1950, 19, 409–430.
- Jacobs, J.M.; Adkins, J.N.; Qian, W.-J.; Liu, T.; Shen, Y.; Camp, D.G.; Smith, R.D. Utilizing Human Blood Plasma for Proteomic Biomarker Discovery. J. Proteome Res. 2005, 4, 1073–1085.
- Shen, Y.; Kim, J.; Strittmatter, E.F.; Jacobs, J.M.; Camp Ii, D.G.; Fang, R.; Tolié, N.; Moore, R.J.; Smith, R.D. Characterization of the human blood plasma proteome. Proteomics 2005, 5, 4034–4045.
- Rai, A.J.; Gelfand, C.A.; Haywood, B.C.; Warunek, D.J.; Yi, J.; Schuchard, M.D.; Mehigh, R.J.; Cockrill, S.L.; Scott, G.B.; Tammen, H.; et al. HUPO Plasma Proteome Project specimen collection and handling: Towards the standardization of parameters for plasma proteome samples. Proteomics 2005, 5, 3262–3277.
- Tuck, M.K.; Chan, D.W.; Chia, D.; Godwin, A.K.; Grizzle, W.E.; Krueger, K.E.; Rom, W.; Sanda, M.; Sorbara, L.; Stass, S.; et al. Standard Operating Procedures for Serum and Plasma Collection: Early Detection Research Network Consensus Statement Standard Operating Procedure Integration Working Group. J. Proteome Res. 2009, 8, 113–117.
- Hampel, H.; O’Bryant, S.E.; Molinuevo, J.L.; Zetterberg, H.; Masters, C.L.; Lista, S.; Kiddle, S.J.; Batrla, R.; Blennow, K. Blood-based biomarkers for Alzheimer disease: Mapping the road to the clinic. Nat. Rev. Neurol. 2018, 14, 639–652.
- Zetterberg, H. Blood-based biomarkers for Alzheimer’s disease-An update. J. Neurosci. Methods 2019, 319, 2–6.
- Milà-Alomà, M.; Suárez-Calvet, M.; Molinuevo, J.L. Latest advances in cerebrospinal fluid and blood biomarkers of Alzheimer’s disease. Ther. Adv. Neurol. Disord. 2019, 12, 1756286419888819.
- Montagne, A.; Barnes, S.R.; Sweeney, M.D.; Halliday, M.R.; Sagare, A.P.; Zhao, Z.; Toga, A.W.; Jacobs, R.E.; Liu, C.Y.; Amezcua, L.; et al. Blood-brain barrier breakdown in the aging human hippocampus. Neuron 2015, 85, 296–302.
- Zetterberg, H.; Blennow, K. From Cerebrospinal Fluid to Blood: The Third Wave of Fluid Biomarkers for Alzheimer’s Disease. J. Alzheimers Dis. 2018, 64, S271–S279.
- Altuna-Azkargorta, M.; Mendioroz-Iriarte, M. Blood biomarkers in Alzheimer’s disease. Neurologia 2021, 36, 704–710.
- Balasa, R.; Barcutean, L.; Balasa, A.; Motataianu, A.; Roman-Filip, C.; Manu, D. The action of TH17 cells on blood brain barrier in multiple sclerosis and experimental autoimmune encephalomyelitis. Hum. Immunol. 2020, 81, 237–243.
- Murphy, C. Olfactory and other sensory impairments in Alzheimer disease. Nat. Rev. Neurol. 2019, 15, 11–24.
- Waldton, S. Clinical observations of impaired cranial nerve function in senile dementia. Acta Psychiatr. Scand. 1974, 50, 539–547.
- Doty, R.L.; Reyes, P.F.; Gregor, T. Presence of both odor identification and detection deficits in Alzheimer’s disease. Brain Res. Bull. 1987, 18, 597–600.
- Doty, R.L.; Hawkes, C.H.; Good, K.P.; Duda, J.E. Odor Perception and Neuropathology in Neurodegenerative Diseases and Schizophrenia. In Handbook of Olfaction and Gustation, 3rd ed.; Wiley Online Books, Doty, R.L., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2015; pp. 403–452.
- Son, G.; Jahanshahi, A.; Yoo, S.J.; Boonstra, J.T.; Hopkins, D.A.; Steinbusch, H.W.M.; Moon, C. Olfactory neuropathology in Alzheimer’s disease: A sign of ongoing neurodegeneration. BMB Rep. 2021, 54, 295–304.
- Kim, J.Y.; Rasheed, A.; Yoo, S.J.; Kim, S.Y.; Cho, B.; Son, G.; Yu, S.W.; Chang, K.A.; Suh, Y.H.; Moon, C. Distinct amyloid precursor protein processing machineries of the olfactory system. Biochem. Biophys. Res. Commun. 2018, 495, 533–538.
- Wesson, D.W.; Levy, E.; Nixon, R.A.; Wilson, D.A. Olfactory dysfunction correlates with amyloid-beta burden in an Alzheimer’s disease mouse model. J. Neurosci. 2010, 30, 505–514.
- Yoo, S.J.; Lee, J.H.; Kim, S.Y.; Son, G.; Kim, J.Y.; Cho, B.; Yu, S.W.; Chang, K.A.; Suh, Y.H.; Moon, C. Differential spatial expression of peripheral olfactory neuron-derived BACE1 induces olfactory impairment by region-specific accumulation of beta-amyloid oligomer. Cell Death Dis. 2017, 8, e2977.
- Son, G.; Steinbusch, H.W.; López-Iglesias, C.; Moon, C.; Jahanshahi, A. Severe histomorphological alterations in post-mortem olfactory glomeruli in Alzheimer’s disease. Brain Pathol. 2022, 32, e13033.
- Braak, H.; Braak, E. Demonstration of amyloid deposits and neurofibrillary changes in whole brain sections. Brain Pathol. 1991, 1, 213–216.
- Masurkar, A.V.; Devanand, D.P. Olfactory Dysfunction in the Elderly: Basic Circuitry and Alterations with Normal Aging and Alzheimer’s Disease. Curr. Geriatr. Rep. 2014, 3, 91–100.
- Attems, J.; Lintner, F.; Jellinger, K.A. Olfactory involvement in aging and Alzheimer’s disease: An autopsy study. J. Alzheimers Dis. 2005, 7, 149–157.
- Crino, P.B.; Martin, J.A.; Hill, W.D.; Greenberg, B.; Lee, V.M.; Trojanowski, J.Q. Beta-Amyloid peptide and amyloid precursor proteins in olfactory mucosa of patients with Alzheimer’s disease, Parkinson’s disease, and Down syndrome. Ann. Otol. Rhinol. Laryngol. 1995, 104, 655–661.
- Hock, C.; Golombowski, S.; Muller-Spahn, F.; Peschel, O.; Riederer, A.; Probst, A.; Mandelkow, E.; Unger, J. Histological markers in nasal mucosa of patients with Alzheimer’s disease. Eur. Neurol. 1998, 40, 31–36.
- Attems, J.; Walker, L.; Jellinger, K.A. Olfactory bulb involvement in neurodegenerative diseases. Acta Neuropathol. 2014, 127, 459–475.
- Franks, K.H.; Chuah, M.I.; King, A.E.; Vickers, J.C. Connectivity of Pathology: The Olfactory System as a Model for Network-Driven Mechanisms of Alzheimer’s Disease Pathogenesis. Front. Aging Neurosci. 2015, 7, 234.
- Struble, R.G.; Clark, H.B. Olfactory bulb lesions in Alzheimer’s disease. Neurobiol. Aging 1992, 13, 469–473.
- Arnold, S.E.; Lee, E.B.; Moberg, P.J.; Stutzbach, L.; Kazi, H.; Han, L.Y.; Lee, V.M.; Trojanowski, J.Q. Olfactory epithelium amyloid-beta and paired helical filament-tau pathology in Alzheimer disease. Ann. Neurol. 2010, 67, 462–469.
- Ayala-Grosso, C.A.; Pieruzzini, R.; Diaz-Solano, D.; Wittig, O.; Abrante, L.; Vargas, L.; Cardier, J. Amyloid-aβ Peptide in olfactory mucosa and mesenchymal stromal cells of mild cognitive impairment and Alzheimer’s disease patients. Brain Pathol. 2015, 25, 136–145.
- Liu, Z.; Kameshima, N.; Nanjo, T.; Shiino, A.; Kato, T.; Shimizu, S.; Shimizu, T.; Tanaka, S.; Miura, K.; Tooyama, I. Development of a High-Sensitivity Method for the Measurement of Human Nasal Aβ42, Tau, and Phosphorylated Tau. J. Alzheimers Dis. 2018, 62, 737–744.
- Yoo, S.J.; Son, G.; Bae, J.; Kim, S.Y.; Yoo, Y.K.; Park, D.; Baek, S.Y.; Chang, K.A.; Suh, Y.H.; Lee, Y.B.; et al. Longitudinal profiling of oligomeric Aβ in human nasal discharge reflecting cognitive decline in probable Alzheimer’s disease. Sci. Rep. 2020, 10, 11234.
- Kim, Y.H.; Lee, S.M.; Cho, S.; Kang, J.H.; Minn, Y.K.; Park, H.; Choi, S.H. Amyloid beta in nasal secretions may be a potential biomarker of Alzheimer’s disease. Sci. Rep. 2019, 9, 4966.
- Lee, J.H.; Goedert, M.; Hill, W.D.; Lee, V.M.; Trojanowski, J.Q. Tau proteins are abnormally expressed in olfactory epithelium of Alzheimer patients and developmentally regulated in human fetal spinal cord. Exp. Neurol. 1993, 121, 93–105.
- Attems, J.; Jellinger, K.A. Olfactory tau pathology in Alzheimer disease and mild cognitive impairment. Clin. Neuropathol. 2006, 25, 265–271.
- Passali, G.C.; Politi, L.; Crisanti, A.; Loglisci, M.; Anzivino, R.; Passali, D. Tau Protein Detection in Anosmic Alzheimer’s Disease Patient’s Nasal Secretions. Chemosens. Percept. 2015, 8, 201–206.
- Moon, J.; Lee, S.T.; Kong, I.G.; Byun, J.I.; Sunwoo, J.S.; Shin, J.W.; Shim, J.Y.; Park, J.H.; Jeon, D.; Jung, K.H.; et al. Early diagnosis of Alzheimer’s disease from elevated olfactory mucosal miR-206 level. Sci. Rep. 2016, 6, 20364.
- Yoshikawa, K.; Wang, H.; Jaen, C.; Haneoka, M.; Saito, N.; Nakamura, J.; Adappa, N.D.; Cohen, N.A.; Dalton, P. The human olfactory cleft mucus proteome and its age-related changes. Sci. Rep. 2018, 8, 17170.
- Zhou, L.; Zhao, S.Z.; Koh, S.K.; Chen, L.; Vaz, C.; Tanavde, V.; Li, X.R.; Beuerman, R.W. In-depth analysis of the human tear proteome. J. Proteom. 2012, 75, 3877–3885.
- Herber, S.; Grus, F.H.; Sabuncuo, P.; Augustin, A.J. Two-dimensional analysis of tear protein patterns of diabetic patients. Electrophoresis 2001, 22, 1838–1844.
- Comoglu, S.S.; Guven, H.; Acar, M.; Ozturk, G.; Kocer, B. Tear levels of tumor necrosis factor-alpha in patients with Parkinson’s disease. Neurosci. Lett. 2013, 553, 63–67.
- Gijs, M.; Nuijts, R.M.; Ramakers, I.; Verhey, F.; Webers, C.A.B. Differences in tear protein biomarkers between patients with Alzheimer’s disease and controls. Investig. Ophth. Vis. Sci. 2019, 60, 1744.
- Gijs, M.; Ramakers, I.H.G.B.; Visser, P.J.; Verhey, F.R.J.; van de Waarenburg, M.P.H.; Schalkwijk, C.G.; Nuijts, R.M.M.A.; Webers, C.A.B. Association of tear fluid amyloid and tau levels with disease severity and neurodegeneration. Sci. Rep. 2021, 11, 22675.
- Wang, Y.R.; Chuang, H.C.; Tripathi, A.; Wang, Y.L.; Ko, M.L.; Chuang, C.C.; Chen, J.C. High-Sensitivity and Trace-Amount Specimen Electrochemical Sensors for Exploring the Levels of beta-Amyloid in Human Blood and Tears. Anal. Chem. 2021, 93, 8099–8106.
- Kallo, G.; Emri, M.; Varga, Z.; Ujhelyi, B.; Tozser, J.; Csutak, A.; Csosz, E. Changes in the Chemical Barrier Composition of Tears in Alzheimer’s Disease Reveal Potential Tear Diagnostic Biomarkers. PLoS ONE 2016, 11, e0158000.
- Kenny, A.; Jimenez-Mateos, E.M.; Zea-Sevilla, M.A.; Rabano, A.; Gili-Manzanaro, P.; Prehn, J.H.M.; Henshall, D.C.; Avila, J.; Engel, T.; Hernandez, F. Proteins and microRNAs are differentially expressed in tear fluid from patients with Alzheimer’s disease. Sci. Rep. 2019, 9, 15437.
- Schenkels, L.C.; Veerman, E.C.; Nieuw Amerongen, A.V. Biochemical composition of human saliva in relation to other mucosal fluids. Crit. Rev. Oral Biol. Med. 1995, 6, 161–175.
- Lamy, E.; Mau, M. Saliva proteomics as an emerging, non-invasive tool to study livestock physiology, nutrition and diseases. J. Proteom. 2012, 75, 4251–4258.
- Vitorino, R.; Guedes, S.; Manadas, B.; Ferreira, R.; Amado, F. Toward a standardized saliva proteome analysis methodology. J. Proteom. 2012, 75, 5140–5165.
- Teresi, L.M.; Kolin, E.; Lufkin, R.B.; Hanafee, W.N. MR imaging of the intraparotid facial nerve: Normal anatomy and pathology. AJR Am. J. Roentgenol. 1987, 148, 995–1000.
- Shi, M.; Sui, Y.T.; Peskind, E.R.; Li, G.; Hwang, H.; Devic, I.; Ginghina, C.; Edgar, J.S.; Pan, C.; Goodlett, D.R.; et al. Salivary Tau Species are Potential Biomarkers of Alzheimer’s Disease. J. Alzheimers Dis. 2011, 27, 299–305.
- Rohleder, N.; Nater, U.M. Determinants of salivary alpha-amylase in humans and methodological considerations. Psychoneuroendocrinology 2009, 34, 469–485.
- Bermejo-Pareja, F.; Antequera, D.; Vargas, T.; Molina, J.A.; Carro, E. Saliva levels of Abeta1-42 as potential biomarker of Alzheimer’s disease: A pilot study. BMC Neurol. 2010, 10, 108.
- Lee, M.; Guo, J.P.; Kennedy, K.; McGeer, E.G.; McGeer, P.L. A Method for Diagnosing Alzheimer’s Disease Based on Salivary Amyloid-beta Protein 42 Levels. J. Alzheimers Dis. 2017, 55, 1175–1182.
- Sabbagh, M.N.; Shi, J.; Lee, M.; Arnold, L.; Al-Hasan, Y.; Heim, J.; McGeer, P. Salivary beta amyloid protein levels are detectable and differentiate patients with Alzheimer’s disease dementia from normal controls: Preliminary findings. BMC Neurol. 2018, 18, 155.
- McGeer, P.L.; Guo, J.P.; Lee, M.; Kennedy, K.; McGeer, E.G. Alzheimer’s Disease Can Be Spared by Nonsteroidal Anti-Inflammatory Drugs. J. Alzheimers Dis. 2018, 62, 1219–1222.
- Kim, C.B.; Choi, Y.Y.; Song, W.K.; Song, K.B. Antibody-based magnetic nanoparticle immunoassay for quantification of Alzheimer’s disease pathogenic factor. J. Biomed. Opt. 2014, 19, 051205.
- Tvarijonaviciute, A.; Zamora, C.; Ceron, J.J.; Bravo-Cantero, A.F.; Pardo-Marin, L.; Valverde, S.; Lopez-Jornet, P. Salivary biomarkers in Alzheimer’s disease. Clin. Oral Investig. 2020, 24, 3437–3444.
- Pekeles, H.; Qureshi, H.Y.; Paudel, H.K.; Schipper, H.M.; Gornistky, M.; Chertkow, H. Development and validation of a salivary tau biomarker in Alzheimer’s disease. Alzheimers Dement. 2019, 11, 53–60.
- Ashton, N.J.; Ide, M.; Scholl, M.; Blennow, K.; Lovestone, S.; Hye, A.; Zetterberg, H. No association of salivary total tau concentration with Alzheimer’s disease. Neurobiol. Aging 2018, 70, 125–127.
- Kruzel, M.L.; Zimecki, M.; Actor, J.K. Lactoferrin in a Context of Inflammation-Induced Pathology. Front. Immunol. 2017, 8, 1438.
- Mayeur, S.; Spahis, S.; Pouliot, Y.; Levy, E. Lactoferrin, a Pleiotropic Protein in Health and Disease. Antioxid. Redox Signal 2016, 24, 813–836.
- Carro, E.; Bartolomé, F.; Bermejo-Pareja, F.; Villarejo-Galende, A.; Molina, J.A.; Ortiz, P.; Calero, M.; Rabano, A.; Cantero, J.L.; Orive, G. Early diagnosis of mild cognitive impairment and Alzheimer’s disease based on salivary lactoferrin. Alzheimers Dement. 2017, 8, 131–138.
- Gonzalez-Sanchez, M.; Bartolome, F.; Antequera, D.; Puertas-Martin, V.; Gonzalez, P.; Gomez-Grande, A.; Llamas-Velasco, S.; Herrero-San Martin, A.; Perez-Martinez, D.; Villarejo-Galende, A.; et al. Decreased salivary lactoferrin levels are specific to Alzheimer’s disease. EBioMedicine 2020, 57, 102834.
- Whitehouse, P.J.; Price, D.L.; Clark, A.W.; Coyle, J.T.; DeLong, M.R. Alzheimer disease: Evidence for selective loss of cholinergic neurons in the nucleus basalis. Ann. Neurol. 1981, 10, 122–126.
- Jann, M.W. Rivastigmine, a new-generation cholinesterase inhibitor for the treatment of Alzheimer’s disease. Pharmacotherapy 2000, 20, 1–12.
- Rinne, J.O.; Kaasinen, V.; Jarvenpaa, T.; Nagren, K.; Roivainen, A.; Yu, M.; Oikonen, V.; Kurki, T. Brain acetylcholinesterase activity in mild cognitive impairment and early Alzheimer’s disease. J. Neurol. Neurosurg. Psychiatry 2003, 74, 113–115.
- Sayer, R.; Law, E.; Connelly, P.J.; Breen, K.C. Association of a salivary acetylcholinesterase with Alzheimer’s disease and response to cholinesterase inhibitors. Clin. Biochem. 2004, 37, 98–104.
- Bakhtiari, S.; Moghadam, N.B.; Ehsani, M.; Mortazavi, H.; Sabour, S.; Bakhshi, M. Can Salivary Acetylcholinesterase be a Diagnostic Biomarker for Alzheimer? J. Clin. Diagn. Res. 2017, 11, ZC58–ZC60.
- Boston, P.F.; Gopalkaje, K.; Manning, L.; Middleton, L.; Loxley, M. Developing a simple laboratory test for Alzheimer’s disease: Measuring acetylcholinesterase in saliva—A pilot study. Int. J. Geriatr. Psychiatry 2008, 23, 439–440.
- Suzuki, Y.J.; Carini, M.; Butterfield, D.A. Protein Carbonylation. Antioxid Redox. Signal 2009, 12, 323–325.
- Aksenov, M.Y.; Aksenova, M.V.; Butterfield, D.A.; Geddes, J.W.; Markesbery, W.R. Protein oxidation in the brain in Alzheimer’s disease. Neuroscience 2001, 103, 373–383.
- Su, H.; Gornitsky, M.; Geng, G.; Velly, A.M.; Chertkow, H.; Schipper, H.M. Diurnal variations in salivary protein carbonyl levels in normal and cognitively impaired human subjects. Age 2008, 30, 1–9.
- Yilmaz, A.; Geddes, T.; Han, B.; Bahado-Singh, R.O.; Wilson, G.D.; Imam, K.; Maddens, M.; Graham, S.F. Diagnostic Biomarkers of Alzheimer’s Disease as Identified in Saliva using 1H NMR-Based Metabolomics. J. Alzheimers Dis. 2017, 58, 355–359.
- Wu, J.Q.; Gao, Y.H. Physiological conditions can be reflected in human urine proteome and metabolome. Expert Rev. Proteomic 2015, 12, 623–636.
- Haubitz, M.; Wittke, S.; Weissinger, E.M.; Walden, M.; Rupprecht, H.D.; Floege, J.; Haller, H.; Mischak, H. Urine protein patterns can serve as diagnostic tools in patients with IgA nephropathy. Kidney Int. 2005, 67, 2313–2320.
- Nam, D.; Lee, J.Y.; Lee, M.; Kim, J.; Seol, W.; Son, I.; Ho, D.H. Detection and Assessment of alpha-Synuclein Oligomers in the Urine of Parkinson’s Disease Patients. J. Parkinson’s Dis. 2020, 10, 981–991.
- Wang, C.; Cui, Y.J.; Yang, J.C.; Zhang, J.R.; Yuan, D.C.; Wei, Y.; Li, Y.M.; Duo, Y.L.; Li, S.J.; Zhu, W.Y.; et al. Combining serum and urine biomarkers in the early diagnosis of mild cognitive impairment that evolves into Alzheimer’s disease in patients with the apolipoprotein E epsilon 4 genotype. Biomarkers 2015, 20, 84–88.
- Luan, H.; Liu, L.F.; Meng, N.; Tang, Z.; Chua, K.K.; Chen, L.L.; Song, J.X.; Mok, V.C.; Xie, L.X.; Li, M.; et al. LC-MS-based urinary metabolite signatures in idiopathic Parkinson’s disease. J. Proteome Res. 2015, 14, 467–478.
- Mischak, H.; Kolch, W.; Aivaliotis, M.; Bouyssié, D.; Court, M.; Dihazi, H.; Dihazi, G.H.; Franke, J.; Garin, J.; Gonzalez de Peredo, A.; et al. Comprehensive human urine standards for comparability and standardization in clinical proteome analysis. Proteom.-Clin. Appl. 2010, 4, 464–478.
- Yamamoto, T. The 4th Human Kidney and Urine Proteome Project (HKUPP) workshop. 26 September 2009, Toronto, Canada. Proteomics 2010, 10, 2069–2070.
- Yamamoto, T.; Langham, R.G.; Ronco, P.; Knepper, M.A.; Thongboonkerd, V. Towards standard protocols and guidelines for urine proteomics: A report on the Human Kidney and Urine Proteome Project (HKUPP) symposium and workshop, 6 October 2007, Seoul, Korea and 1 November 2007, San Francisco, CA, USA. Proteomics 2008, 8, 2156–2159.
- Zürbig, P.; Dihazi, H.; Metzger, J.; Thongboonkerd, V.; Vlahou, A. Urine proteomics in kidney and urogenital diseases: Moving towards clinical applications. Proteom.-Clin. Appl. 2011, 5, 256–268.
- Takata, M.; Nakashima, M.; Takehara, T.; Baba, H.; Machida, K.; Akitake, Y.; Ono, K.; Hosokawa, M.; Takahashi, M. Detection of amyloid beta protein in the urine of Alzheimer’s disease patients and healthy individuals. Neurosci. Lett. 2008, 435, 126–130.
- Wongta, A.; Hongsibsong, S.; Chantara, S.; Pattarawarapan, M.; Sapbamrer, R.; Sringarm, K.; Xu, Z.L.; Wang, H. Development of an Immunoassay for the Detection of Amyloid Beta 1-42 and Its Application in Urine Samples. J. Immunol. Res. 2020, 2020, 8821181.
- Yao, F.; Hong, X.; Li, S.; Zhang, Y.; Zhao, Q.; Du, W.; Wang, Y.; Ni, J. Urine-Based Biomarkers for Alzheimer’s Disease Identified Through Coupling Computational and Experimental Methods. J. Alzheimers Dis. 2018, 65, 421–431.
- Ku, B.D.; Kim, H.; Kim, Y.K.; Ryu, H.U. Comparison of Urinary Alzheimer-Associated Neural Thread Protein (AD7c-NTP) Levels Between Patients With Amnestic and Nonamnestic Mild Cognitive Impairment. Am. J. Alzheimers Dis. 2020, 35, 1533317519880369.
- Watanabe, Y.; Hirao, Y.; Kasuga, K.; Tokutake, T.; Semizu, Y.; Kitamura, K.; Ikeuchi, T.; Nakamura, K.; Yamamoto, T. Molecular Network Analysis of the Urinary Proteome of Alzheimer’s Disease Patients. Dement. Geriatr. Cogn. Dis. Extra 2019, 9, 53–65.
- Watanabe, Y.; Hirao, Y.; Kasuga, K.; Tokutake, T.; Kitamura, K.; Niida, S.; Ikeuchi, T.; Nakamura, K.; Yamamoto, T. Urinary Apolipoprotein C3 Is a Potential Biomarker for Alzheimer’s Disease. Dement. Geriatr. Cogn. Dis. Extra 2020, 10, 94–104.
- Nicholson, J.K.; Holmes, E.; Kinross, J.; Burcelin, R.; Gibson, G.; Jia, W.; Pettersson, S. Host-gut microbiota metabolic interactions. Science 2012, 336, 1262–1267.
- Vogt, N.M.; Kerby, R.L.; Dill-McFarland, K.A.; Harding, S.J.; Merluzzi, A.P.; Johnson, S.C.; Carlsson, C.M.; Asthana, S.; Zetterberg, H.; Blennow, K.; et al. Gut microbiome alterations in Alzheimer’s disease. Sci. Rep. 2017, 7, 13537.
- Giau, V.V.; Wu, S.Y.; Jamerlan, A.; An, S.S.A.; Kim, S.Y.; Hulme, J. Gut Microbiota and Their Neuroinflammatory Implications in Alzheimer’s Disease. Nutrients 2018, 10, 1765.
- Kurbatova, N.; Garg, M.; Whiley, L.; Chekmeneva, E.; Jimenez, B.; Gomez-Romero, M.; Pearce, J.; Kimhofer, T.; D’Hondt, E.; Soininen, H.; et al. Urinary metabolic phenotyping for Alzheimer’s disease. Sci. Rep. 2020, 10, 21745.
- Pena-Bautista, C.; Vigor, C.; Galano, J.M.; Oger, C.; Durand, T.; Ferrer, I.; Cuevas, A.; Lopez-Cuevas, R.; Baquero, M.; Lopez-Nogueroles, M.; et al. New screening approach for Alzheimer’s disease risk assessment from urine lipid peroxidation compounds. Sci. Rep. 2019, 9, 14244.
- Mall, C.; Rocke, D.M.; Durbin-Johnson, B.; Weiss, R.H. Stability of miRNA in human urine supports its biomarker potential. Biomark. Med. 2013, 7, 623–631.
- Seol, W.; Kim, H.; Son, I. Urinary Biomarkers for Neurodegenerative Diseases. Exp. Neurobiol. 2020, 29, 325–333.
- Cheng, L.; Sun, X.; Scicluna, B.J.; Coleman, B.M.; Hill, A.F. Characterization and deep sequencing analysis of exosomal and non-exosomal miRNA in human urine. Kidney Int. 2014, 86, 433–444.
