Microalgal biomass and metabolites can be used as a renewable source of nutrition, pharmaceuticals and energy to maintain or improve the quality of human life. Microalgae’s high volumetric productivity and low impact on the environment make them a promising raw material in terms of both ecology and economics. To optimize biotechnological processes with microalgae, improving the productivity and robustness of the cell factories is a major step towards economically viable bioprocesses. The success of a random mutagenesis approach using microalgae is determined by multiple factors involving the treatment of the cells before, during and after the mutagenesis procedure. Using photosynthetic microalgae, the supply of light quality and quantity, as well as the supply of carbon and nitrogen, are the most important factors. Besides the environmental conditions, the type of mutagen, its concentration and exposure time are among the main factors affecting the mutation result.
- random mutagenesis
- algae
- mutagens
- strain development
- microalgal biotechnology
1. Physical Mutagens in Microalgal Biotechnology
1.1. Ultraviolet Light
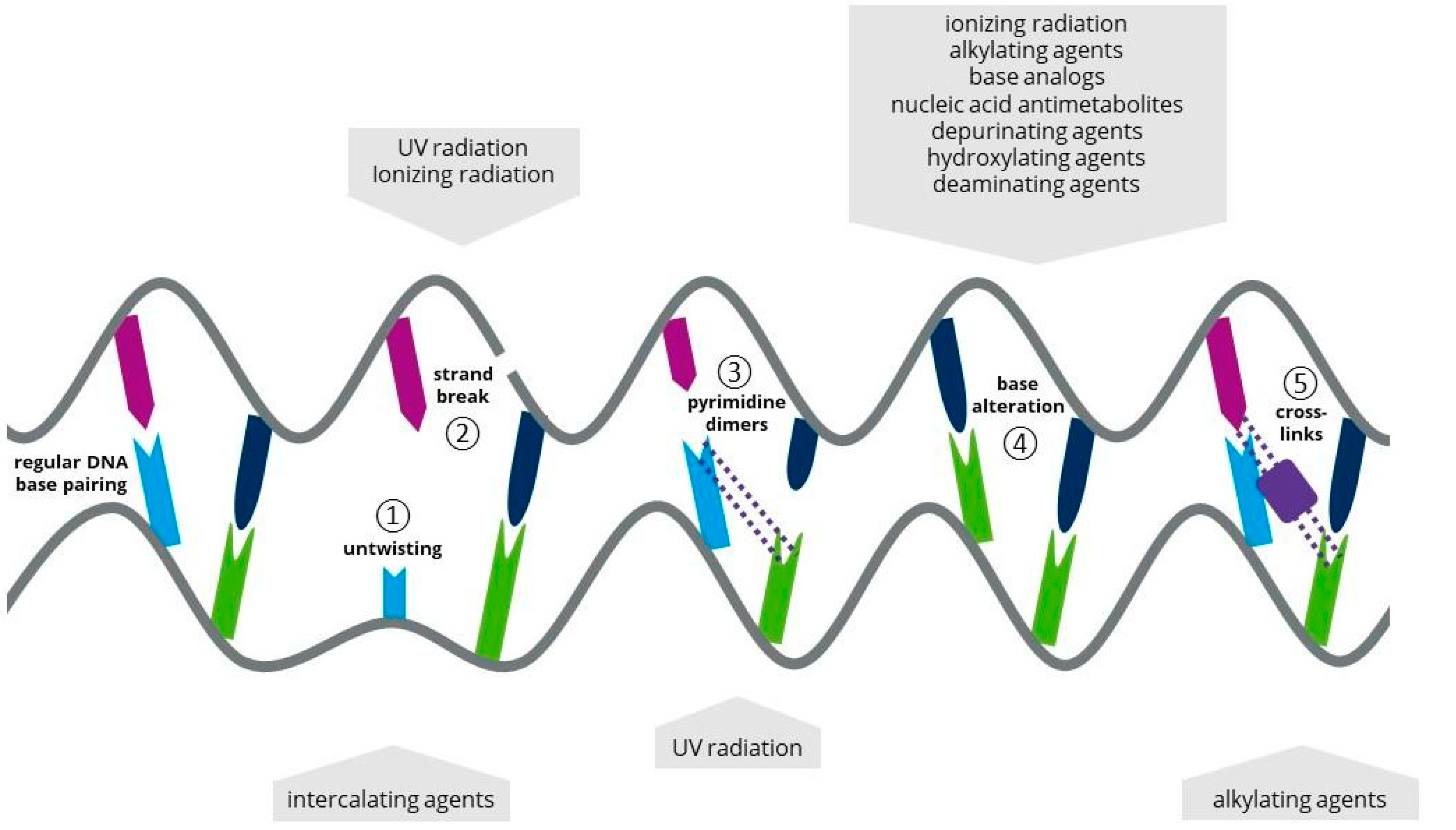
1.2. Ionizing Radiation
1.3. Atmospheric and Room Temperature Plasma
1.4. Laser Radiation
| Mutagen | Method, Exposure Time, Source, Distance, Recovery Time | Reference Microalgae | Mutation Results | References | ||
|---|---|---|---|---|---|---|
| Mutated trait |
WT * | M ** | ||||
| UV | UV 18 W, for 13 min, 15 cm, 24 h darkness | Chlorella vulgaris Y-019 |
neutral lipid accumulation [g/g dry wt] |
0.11 | 0.26 | [24] |
| UV-C | UV-C 253.7 nm, 30-W, 3–30 min, 9 cm, 24 h darkness | Chlorella sp. | protein content [g/L] | 0.0242 | 0.0688 | [25] |
| UV-C 254 nm 1.4 mW/cm2 for 60 s, 15 cm, 16 h darkness | Chlorella vulgaris | fatty acids 16:0;18:0, 20:0 [% of total fatty acids] | 27.9; 3.9; 11.9 | 47.4; 5.9; 19.9 | [26] | |
| UV-C 254 nm, 15 W, (Vilber–Lourmat, France), for 30–180 s, 5 cm, 24 h darkness | natural isolates of photosynthetic microorganism | lipid content though Nile red autofluorescence; with fluorescence emission | 35; 1081 | 983; 89,770 | [27] | |
| UV-C 40,000 μJ/cm, 254 nm, overnight darkness | Scenedesmus obliquus | trans-fatty acid productivity [g/(L·d)] |
0.095 | 0.112 | [28] | |
| UV-C 254 nm 340 mW cm2, for 3–32 min, 13.5 cm, 24 h darkness |
Isochrysis affinis galbana | total fatty acid [g/g dry wt] |
0.262 | 0.409 | [29] | |
| UV-C, for 1–10 min, 40 cm, overnight darkness | Chlorella vulgaris | lipid content [g/g] | 0.58 | 0.75 | [30] | |
| Gamma irradiation | 10 doses of irradiation 50–7000 kGy, 60Co gamma ray irradiator, room temperature |
Scenedesmus sp. | lipid productivity [g/L·d] |
0.0648 | 0.097 | [31] |
| ARTP | He RF power 100 W, plasma temperature 25–35 °C, for 20; 40; 60 and 80 s, 2 mm | Spirulina platensis | Carbohydrates productivity [g/L·d] |
0.0157 | 0.026 | [15] |
| He RF power 100 W, plasma temperature 25–35 °C, 20–60 s, 2 mm | Chlamydomonas reinhardtii | H2 production [mL/L] | ~16.1 | 84.1 | [32] | |
| He RF power 150 W, for 100 s | Crypthecodinium cohnii | biomass concentration [g dry wt/L] |
3.60 | 4.24 | [33] | |
| Heavy ion beam | 12 C6+ ion beam 31 keVµm−1 160 Gy, | Nannochloropsis oceanica | lipid productivity [g/L·d] | 0.211 | 0.295 | [34] |
| 12 C6+ ion beam, 90 Gy | Desmodesmus sp. | lipid productivity [g/L·d] | 0.247 | 0.298 | [35] | |
| Low-energy ion beam implementation | N+ ion beam chamber pressure 10−2 Pa Dose of implantation 0.3–3.3·1015 ions cm−2 s−1 |
Chlorella pyrenoidosa | lipid productivity [g/ L·d]; Lipid content [g/g dry wt] | 47.7; 0.337 | 64.4; 0.446 | [36] |
| laser radiation | He–Ne laser 808 nm, 6 W, 4 min, 24 h darkness | C. pyrenoidesa | lipid content [g/g dry wt] | 0.354 | 0.780 | [22] |
| Nd:YAG laser 1064 nm, 40 mW 8 min, 24 h darkness | Chlorella vulgaris | lipid content [g/g dry wt] | 0.315 | 0.525 | [22] | |
| Nd:YAG laser 1064 nm, 40 mW 2 min, 24 h darkness | Chlorella pacifica | lipid content [g L−1] |
0.033 | 0.088 | [37] | |
| semiconductor laser 632 nm, 40 mW, 4 min, 24 h darkness |
Chlorella pacifica | lipid content [g L−1] |
0.033 | 0.077 | [37] | |
2. Chemical Mutagens in Microalgal Biotechnology
2.1. Alkylating Agents as a Chemical Mutagen
2.2. Base Analogs (BAs) as a Chemical Mutagen
2.3. Antimetabolites (AMs) as a Chemical Mutagen
2.4. Intercalating Agents (IAs) as a Chemical Mutagen
2.5. Other Approaches for Chemical Mutagenesis
| Mutagen | Mutagen Concentration, Time of Exposure | Reference Microalgae | Mutation Results | References | ||
|---|---|---|---|---|---|---|
| Mutated trait |
WT * | M ** | ||||
| EMS | EMS 0.1–1.2 M for 60 min |
Nannochloropsis sp. | fatty acid methyl esters [g/g of dry wt] | 0.123 | 0.238 | [65] |
| EMS 0.4–1 g/L for 60–120 min |
Haematococcus pluvialis | total carotenoid; Astaxanthin [g/g of dry wt] |
0.02; 0.005 | 0.02; 0.019 |
[66] | |
| EMS 300 mM for 60 min | Chlorella vulgaris | protein content [g/g of dry wt] | 0.353 | 0.455 | [67] | |
| EMS 0.2–0.4 M for 2 h in darkness | Chlorella vulgaris | violaxanthin [mg/L culture] | 1.64 | 5.23 | [68] | |
| EMS 0.1–0.2 M | Phaeodactylum tricornutum | total carotenoids [g/g dry wt] | 0.009 | 0.011 | [69] | |
| EMS 0.2 M for 2 h in the dark |
Dunaliella tertiolecta | Zeaxanthin [μg/106·cells] | 0.131 | 0.359 | [70] | |
| EMS 20–40 µL/mL for 2 h | Chlamydomonas reinhardtii |
fatty acid methyl esters yield [%] | 6.53 | 7.56 | [71] | |
| EMS 0.2 M for 2 h in the dark |
Dunaliella salina |
carotenoid synthesis [Mol Car/Mol Chl] | 0.99 | 1.24 | [72] | |
| EMS 100 μ mol mL−1, for 30 min | Chlorella sp. | lipid content [g/g of dry wt]; productivity [g/(L·d)] | 0.247; 0.1536 | 0.356; 0.2487 | [73] | |
| EMS 0.4 M, for 60 min | Coelastrum sp. | Astaxanthin content [g/L] | 0.0145 | 0.0283 | [74] | |
| EMS + UV | UV + EMS 25 mM for 60 min | Chlorella vulgaris | lipid content [%] | 100 | 167 | [46] |
| UV 5–240 s, 245 nm + EMS 0.24 mol/L for 30 min | Nannochloropsis salina | fatty acid methyl ester [g/g of dry wt] | 0.175 | 0.787 | [75] | |
| MNNG | MNNG 0.1 mM for 60 min | Haematococcus pluvialis | Total carotenoid content [g/L] | ~0.067 | 0.089 | [41] |
| MNNG 5 µg/mL for 60 min | Chlorella sp. | max. growth rate under alkaline conditions [ d−1] | 0.064 | 0.554 | [76] | |
| MNNG 0.02 mol/L for 60 min |
Nannochloropsis oceanica |
Total lipid content [g/g] Lipid productivity [g/(L·d)] |
0.241; 0.0065 | 0.299; 0.0086 | [50] | |
| MNNG 0.1–0.2 M | Phaeodactylum tricornutum | total carotenoids [g/g dry wt] | 0.009 | 0.011 | [69] | |
| MNNG 0.2 mg/mL | Chlorella sorokiniana | Lutein content [g/L] | 0.025 | 0.042 | [44] | |
| MNNG 0.25–0.5 mM | Botryosphaerella sp. | lipid [g dry wt/(m2 day)]; biomass productivity [g dry wt/(m2·day)] | 1.0; 3.2 | 1.9; 5.4 | [45] | |
| NMU | NMU 5 mM for 60–90 min |
Nannochloropsis oculata | Total fatty acid [g/g dry wt] | 0.0634 | 0.0762 | [43] |
| DES + UV | UV 7–11 min 254 nm + DES 0.1–1.5% (V/V) 40 min |
Haematococcus pluvialis | astaxanthin content [mg/L] | ~0.031 | ~0.089 | [42] |
| 5BU | 5BU 1 mM for 48 h | Chlamydomonas reinhardtii | O2 tolerance [%] | 100 | 1400 | [77] |
| 5′FDU | 5′FDU 0.25 and 0.50 mM for 1 week | Chlorella vulgaris | fatty acids 16:0; 18:0; 20:0 [% of total fatty acids] |
27.9; 3.9; 11.9 | 46.9; 5.5; 18.5 | [26] |
| Acriflavin | Acriflavin 2–8 μg/mL for 1–3 d in darkness | Chlamydomonas reinhardtii zyklo | Loss of respiratory rate [nmol O2/(min·107 cells)] through loss of mitochondrial DNA | 23.2 | 3.7 | [64] |
3. Further Approaches in Random Mutagenesis
Recently, combined mutagenesis approaches have generated high interest as results indicated that they have a higher success rate than individual approaches. For instance, Wang et al. [42] applied a two-step random mutagenesis protocol to Haematococcus pluvialis cells using first UV irradiation, then EMS and DES mutagenesis, causing astaxanthin production to increase by a factor of 1.7 compared to the wild strain. Beacham et al. [75] used a reverse protocol for Nannochloropsis salina, starting with exposure to EMS, followed by UV irradiation, yielding a three-fold increase in cellular lipid accumulation. Comparable results were achieved by Sivaramakrishnan and Incharoensakdi [78], who exposed Scenedesmus sp. to UV irradiation in combination with oxidative stress by H2O2.
Other approaches can be used to select desired microalgal cells if the results obtained by random mutagenesis are insufficient. Among them, Adaptive Laboratory Evolution (ALE) is commonly used to adapt the physiology of cells to specific process conditions, such as high temperatures [79]. Its principle is based on natural selection, as presented in the Darwinian Theory, on the laboratory bench [80], and includes extensive cultivation in a specifically designed lab environment so that enhanced phenotypes can be selected after a long period of time [81]. The environmental conditions that can be altered include light irradiation, lack of nutrients, such as nitrogen, osmotic, temperature and oxidative stress [80][82][83]. Connecting the results of ALE with whole genome sequencing and “omics” methods enables gene functions to be discovered easily [81]. However, ALE does not prevent gene instability that might occur more often than in randomly mutated cells [79][82].
Additional environmental factors can be applied on microalgae; for example, Miazek et al. [84] reviewed the use of metals, metalloids and metallic nanoparticles to enhance cell characteristics. Moreover, phytohormones or chemicals acting as metabolic precursors have already been applied to microalgae [85]. A discussion of the methods used in the latter case exceeds the scope of this research.
More recently, a new technique was developed, known as Space Mutation Breeding (SMB). This technique may have direct or indirect effects on the growth and metabolic activities of microalgae, due to the unusual environment of space, characterized by high-energy ionic radiation, space’s magnetic field, ultra-high vacuum and microgravity [86]. The SMB technique provides some advantages, such as the great improvement in species’ qualities in a short time [87]. This was achieved by Chen Zishuo et al. [86], with a seawater Arthrospira platensis mutant, yielding a sugar content 62.26% higher than the wild type.
This entry is adapted from the peer-reviewed paper 10.3390/life12070961
