Epigenetic changes in DNA methylation contribute to the development of many diseases, including cancer. In glioblastoma multiforme, the most prevalent primary brain cancer and an incurable tumor with a median survival time of 15 months, a single epigenetic modification, the methylation of the O6-Methylguanine-DNA Methyltransferase (MGMT) gene, is a valid biomarker for predicting response to therapy with alkylating agents and also, independently, prognosis. More recently, the progress from single gene to whole-genome analysis of DNA methylation has allowed a better subclassification of glioblastomas.
- glioblastoma multiforme
- DNA Methylation
- techniques
- MGMT
- whole-genome methylation profiling
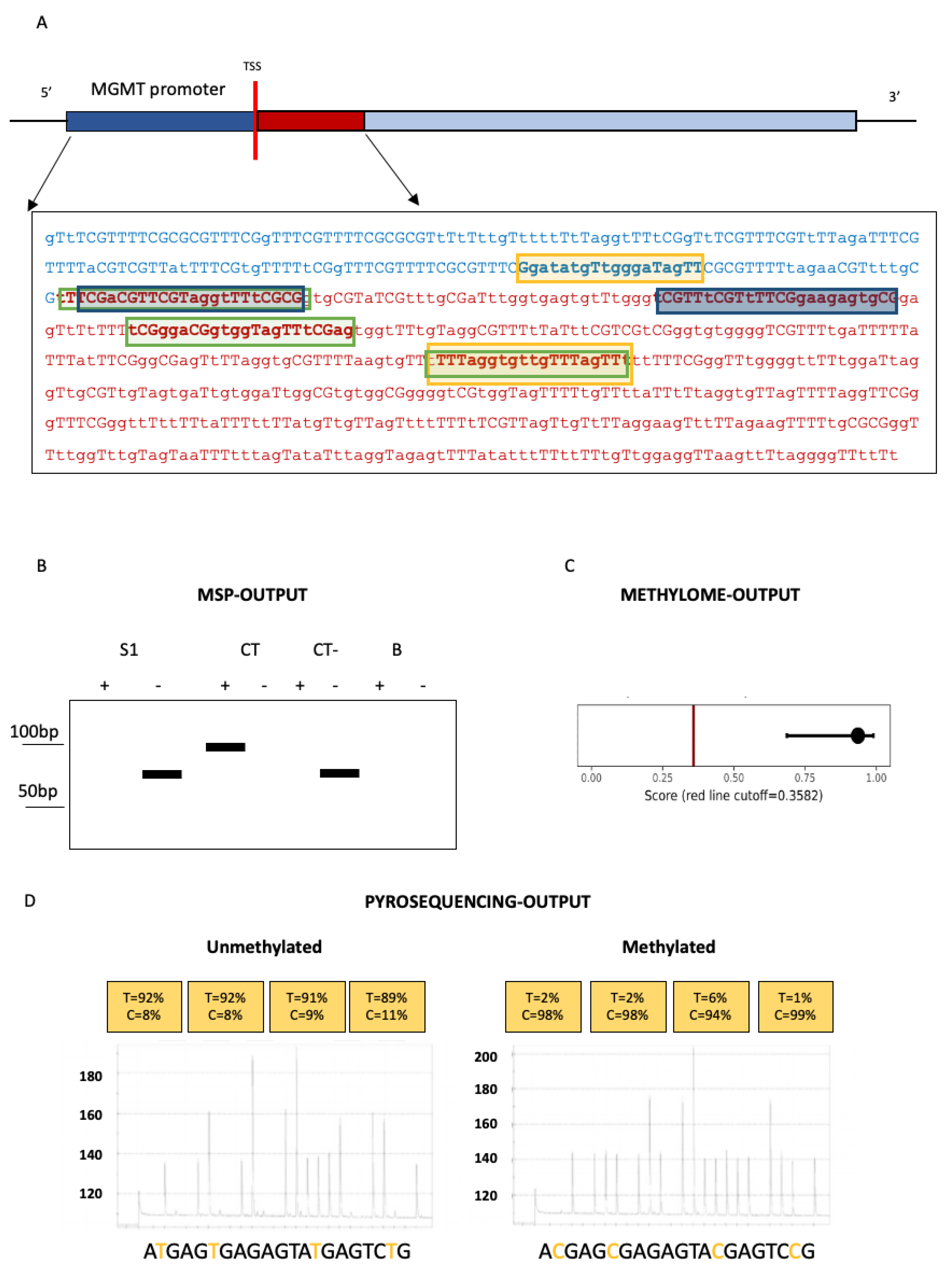
1. Techniques for O6-Methylguanine-DNA Methyltransferase(MGMT) Methylation Assessment in Glioblastomas
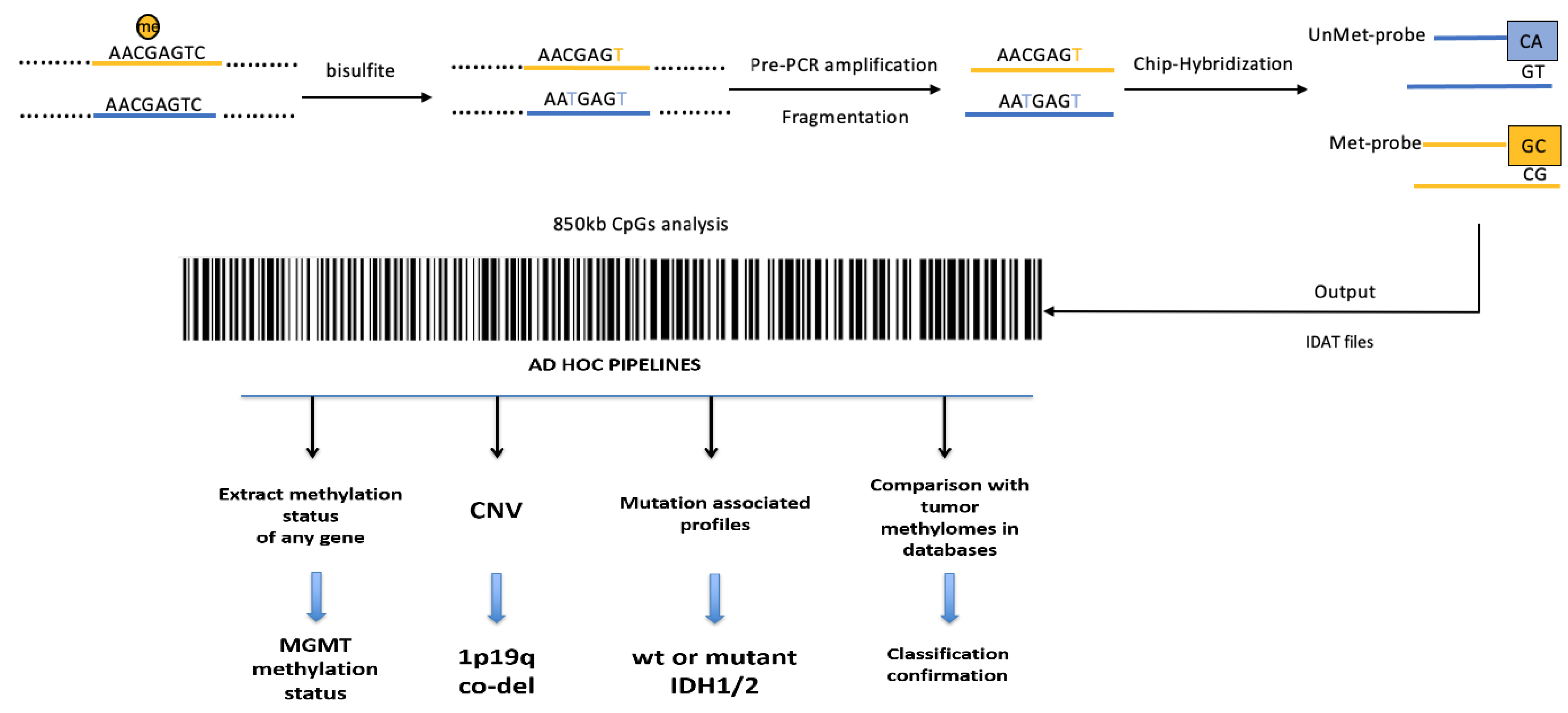
2. Whole-Genome Methylation Profiling (Methylome) of Glioblastomas
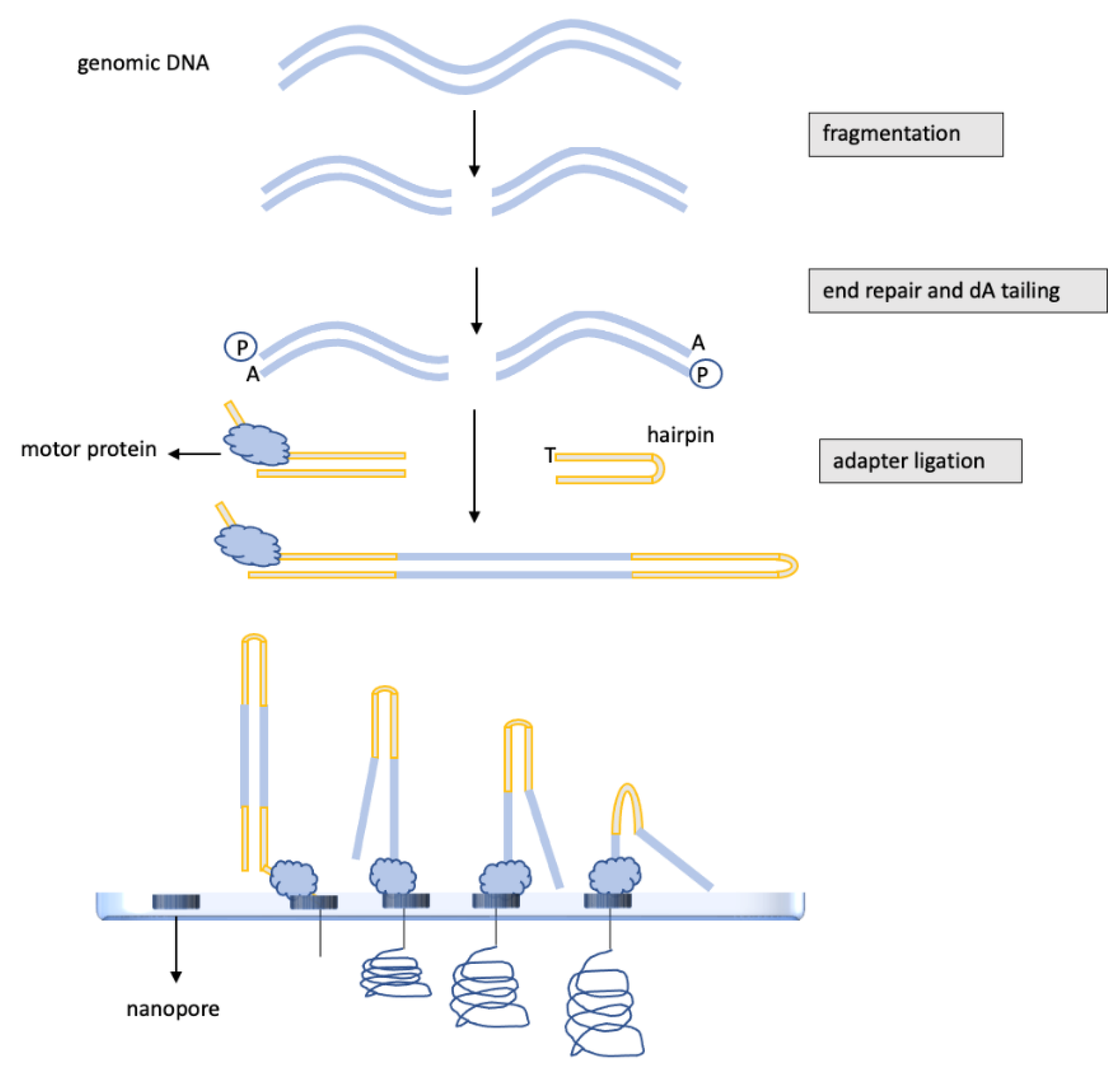
3. DNA Methylation Analysis of Glioblastomas by Nanopore, “Third-Generation” Sequencing
This entry is adapted from the peer-reviewed paper 10.3390/ijms23137148
References
- Philteos, J.; Karmur, B.S.; Mansouri, A. MGMT testing in glioblastomas: Pitfalls and opportunities. Am. J. Clin. Oncol. 2019, 42, 117–122.
- Herman, J.G.; Graff, J.R.; Myöhänen, S.; Nelkin, B.D.; Baylin, S.B. Methylation-specific PCR: A novel PCR assay for methylation status of CpG islands. Proc. Natl. Acad. Sci. USA 1996, 93, 9821–9826.
- Della Monica, R.; Cuomo, M.; Visconti, R.; di Mauro, A.; Buonaiuto, M.; Costabile, D.; De Riso, G.; Di Risi, T.; Guadagno, E.; Tafuto, R.; et al. Evaluation of MGMT gene methylation in neuroendocrine neoplasms. Oncol. Res. 2022, 28, 837–845.
- Hegi, M.E.; Diserens, A.C.; Godard, S.; Dietrich, P.Y.; Regli, L.; Ostermann, S.; Otten, P.; Van Melle, G.; de Tribolet, N.; Stupp, R. Clinical trial substantiates the predictive value of 0-6-methylguanine-DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clin. Cancer Res. 2004, 10, 871–874.
- Esteller, M.; Hamilton, S.R.; Burger, P.C.; Baylin, S.B.; Herman, J.G. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999, 59, 793–797.
- Gilbert, M.R.; Wang, M.; Aldape, K.D.; Stupp, R.; Hegi, M.E.; Jaeckle, K.A.; Armstrong, T.S.; Wefel, J.S.; Won, M.; Blumenthal, D.T.; et al. Dose-dense temozolomide for newly diagnosed glioblastoma: A randomized phase III clinical trial. J. Clin. Oncol. 2013, 31, 4085–4091.
- Vlassenbroeck, I.; Califice, S.; Diserens, A.C.; Migliavacca, E.; Straub, J.; Di Stefano, I.; Moreau, F.; Hamou, M.F.; Renard, I.; Delorenzi, M.; et al. Validation of real-time methylation-specific PCR to determine O6-methylguanine-DNA methyltransferase gene promoter methylation in glioma. J. Mol. Diagn. 2008, 10, 332–337.
- Wojdacz, T.K.; Dobrovic, A. Methylation-sensitive high resolution melting (MS-HRM): A new approach for sensitive and high-throughput assessment of methylation. Nucleic Acids Res. 2007, 35, e41.
- Switzeny, O.J.; Christmann, M.; Renovanz, M.; Giese, A.; Sommer, C.; Kaina, B. MGMT promoter methylation determined by HRM in comparison to MSP and pyrosequencing for predicting high-grade glioma response. Clin. Epigenetics 2016, 8, 49.
- Estival, A.; Sanz, C.; Ramirez, J.L.; Velarde, J.M.; Domenech, M.; Carrato, C.; de Las Peñas, R.; Gil-Gil, M.; Sepúlveda, J.; Armengol, R.; et al. Pyrosequencing versus methylation-specific PCR for assessment of MGMT methylation in tumor and blood samples of glioblastoma patients. Sci. Rep. 2019, 9, 11125.
- Karayan-Tapon, L.; Quillien, V.; Guilhot, J.; Wager, M.; Fromont, G.; Saikali, S.; Etcheverry, A.; Hamlat, A.; Loussouarn, D.; Campion, L.; et al. Prognostic value of O6-methylguanine-DNA methyltransferase status in glioblastoma patients, assessed by five different methods. J. Neuro-Oncology 2010, 97, 311–322.
- Christians, A.; Hartmann, C.; Benner, A.; Meyer, J.; von Deimling, A.; Weller, M.; Wick, W.; Weiler, M. Prognostic value of three different methods of MGMT promoter methylation analysis in a prospective trial on newly diagnosed glioblastoma. PLoS ONE 2012, 7, e33449.
- Quillien, V.; Lavenu, A.; Karayan-Tapon, L.; Carpentier, C.; Labussière, M.; Lesimple, T.; Chinot, O.; Wager, M.; Honnorat, J.; Saikali, S.; et al. Comparative assessment of 5 methods (methylation-specific polymerase chain reaction, MethyLight, pyrosequencing, methylation-sensitive high-resolution melting, and immunohistochemistry) to analyze O6-methylguanine-DNA-methyltranferase in a series of 100 glioblastoma patients. Cancer 2012, 118, 4201–4211.
- Jeuken, J.W.; Cornelissen, S.J.; Vriezen, M.; Dekkers, M.M.; Errami, A.; Sijben, A.; Boots-Sprenger, S.H.; Wesseling, P. MS-MLPA: An attractive alternative laboratory assay for robust, reliable, and semiquantitative detection of MGMT promoter hypermethylation in gliomas. Lab. Investig. 2007, 87, 1055–1065.
- Shah, N.; Lin, B.; Sibenaller, Z.; Ryken, T.; Lee, H.; Yoon, J.G.; Rostad, S.; Foltz, G. Comprehensive analysis of MGMT promoter methylation: Correlation with MGMT expression and clinical response in GBM. PLoS ONE 2011, 6, e16146.
- Trabelsi, S.; Mama, N.; Ladib, M.; Karmeni, N.; Haddaji Mastouri, M.; Chourabi, M.; Mokni, M.; Tlili, K.; Krifa, H.; Yacoubi, M.T.; et al. MGMT methylation assessment in glioblastoma: MS-MLPA versus human methylation 450K beadchip array and immunohistochemistry. Clin. Transl. Oncol. 2016, 18, 391–397.
- Mancarella, D.; Plass, C. Epigenetic signatures in cancer: Proper controls, current challenges and the potential for clinical translation. Genome Med. 2021, 13, 23.
- Orjuela, S.; Menigatti, M.; Schraml, P.; Kambakamba, P.; Robinson, M.D.; Marra, G. The DNA hypermethylation phenotype of colorectal cancer liver metastases resembles that of the primary colorectal cancers. BMC Cancer 2020, 20, 290.
- Modhukur, V.; Sharma, S.; Mondal, M.; Lawarde, A.; Kask, K.; Sharma, R.; Salumets, A. Machine learning approaches to classify primary and metastatic cancers using tissue of origin-based DNA methylation profiles. Cancers 2021, 13, 3768.
- Smit, K.N.; Boers, R.; Vaarwater, J.; Boers, J.; Brands, T.; Mensink, H.; Verdijk, R.M.; van Ijcken, W.F.J.; Gribnau, J.; de Klein, A.; et al. Genome-wide aberrant methylation in primary metastatic UM and their matched metastases. Sci. Rep. 2022, 12, 42.
- Capper, D.; Jones, D.T.W.; Sill, M.; Hovestadt, V.; Schrimpf, D.; Sturm, D.; Koelsche, C.; Sahm, F.; Chavez, L.; Reuss, D.E.; et al. A methylation-based classification of central nervous system tumours. Nature 2018, 555, 469–474.
- Epigenetic Lab—CEINGE, Advanced Biotechnologies. Available online: https://www.ceinge.unina.it/en/epigenetic-lab (accessed on 9 May 2022).
- Patel, A.P.; Tirosh, I.; Trombetta, J.J.; Shalek, A.K.; Gillespie, S.M.; Wakimoto, H.; Cahill, D.P.; Nahed, B.V.; Curry, W.T.; Martuza, R.L.; et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014, 344, 1396–1401.
- Neftel, C.; Laffy, J.; Filbin, M.G.; Hara, T.; Shore, M.E.; Rahme, G.J.; Richman, A.R.; Silverbush, D.; Shaw, M.L.; Hebert, C.M.; et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell 2019, 178, 835–849.
- Wenger, A.; Ferreyra Vega, S.; Kling, T.; Bontell, T.O.; Jakola, A.S.; Carén, H. Intratumor DNA methylation heterogeneity in glioblastoma: Implications for DNA methylation-based classification. Neuro-Oncology 2019, 21, 616–627.
- Parker, N.R.; Hudson, A.L.; Khong, P.; Parkinson, J.F.; Dwight, T.; Ikin, R.J.; Zhu, Y.; Cheng, Z.J.; Vafaee, F.; Chen, J.; et al. Intratumoral heterogeneity identified at the epigenetic, genetic and transcriptional level in glioblastoma. Sci. Rep. 2016, 6, 22477.
- Akgül, S.; Patch, A.M.; D’Souza, R.C.J.; Mukhopadhyay, P.; Nones, K.; Kempe, S.; Kazakoff, S.H.; Jeffree, R.L.; Stringer, B.W.; Pearson, J.V.; et al. Intratumoural heterogeneity underlies distinct therapy responses and treatment resistance in glioblastoma. Cancers 2019, 11, 190.
- Ramasamy, D.; Deva Magendhra Rao, A.K.; Rajkumar, T.; Mani, S. Non-CpG methylation—A key epigenetic modification in cancer. Brief. Funct. Genom. 2021, 20, 304–311.
- Brigliadori, G.; Goffredo, G.; Bartolini, D.; Tosatto, L.; Gurrieri, L.; Mercatali, L.; Ibrahim, T. Influence of intratumor heterogeneity on the predictivity of MGMT gene promoter methylation status in glioblastoma. Front. Oncol. 2020, 10, 533000.
- Chai, R.-C.; Liu, Y.-Q.; Zhang, K.-N.; Wu, F.; Zhao, Z.; Wang, K.-Y.; Jiang, T.; Wang, Y.-Z. A novel analytical model of MGMT methylation pyrosequencing offers improved predictive performance in patients with gliomas. Mod. Pathol. 2019, 32, 4–15.
- Pezone, A.; Tramontano, A.; Scala, G.; Cuomo, M.; Riccio, P.; De Nicola, S.; Porcellini, A.; Chiariotti, L.; Avvedimento, E.V. Tracing and tracking epiallele families in complex DNA populations. NAR Genom. Bioinform. 2020, 2, lqaa096.
- Amarasinghe, S.L.; Su, S.; Dong, X.; Zappia, L.; Ritchie, M.E.; Gouil, Q. Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 2020, 21, 30.
- Patel, A.; Dogan, H.; Payne, A.; Krause, E.; Sievers, P.; Schoebe, N.; Schrimpf, D.; Blume, C.; Stichel, D.; Holmes, N.; et al. Rapid-CNS2: Rapid comprehensive adaptive nanopore-sequencing of CNS tumors, a proof-of-concept study. Acta Neuropathol. 2022, 143, 609–612.
- Gilpatrick, T.; Lee, I.; Graham, J.E.; Raimondeau, E.; Bowen, R.; Heron, A.; Downs, B.; Sukumar, S.; Sedlazeck, F.J.; Timp, W. Targeted nanopore sequencing with Cas9-guided adapter ligation. Nat. Biotechnol. 2020, 38, 433–438.
- Wongsurawat, T.; Jenjaroenpun, P.; De Loose, A.; Alkam, D.; Ussery, D.W.; Nookaew, I.; Leung, Y.K.; Ho, S.M.; Day, J.D.; Rodriguez, A. A novel Cas9-targeted long-read assay for simultaneous detection of IDH1/2 mutations and clinically relevant MGMT methylation in fresh biopsies of diffuse glioma. Acta Neuropathol. Commun. 2020, 8, 87.
- Djirackor, L.; Halldorsson, S.; Niehusmann, P.; Leske, H.; Capper, D.; Kuschel, L.P.; Pahnke, J.; Due-Tønnessen, B.J.; Langmoen, I.A.; Sandberg, C.J.; et al. Intraoperative DNA methylation classification of brain tumors impacts neurosurgical strategy. Neuro-Oncology Adv. 2021, 3, vdab149.
