Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
The rhythmicity of gut microbiota has fundamental implications for host physiology, metabolism, and health. The microbial rhythmicity is affected by many host-derived factors including the host circadian clock. Vice versa, gut microbiota rhythmicity can influence the host’s circadian rhythm.
- diurnal rhythmicity
- gut microbiota
- circadian rhythm
1. Introduction
The rotation of the earth shapes the periodic changes of the environmental cues including light and temperature, which are tightly correlated to the physiology and metabolism of mammals. The circadian clock has thus evolved to adjust to the periodic changes of the environmental cues. The circadian clock, an intrinsic timing system with an approximate 24-h period, orchestrates the physiological functions and activities like eating, sleep-wake, and hormone secretion, and further synchronizes these activities with the changing environment [1]. The circadian timekeeper was initially supposed to only exist in higher organisms, including animals and plants. Bacteria is considered too simple to develop a circadian clock. Nevertheless, evidence illuminates that a diurnal rhythm exists in cyanobacteria involved in photosynthesis and nitrogen fixation [2][3]. Three clock proteins, including KaiA, KaiB, and KaiC, make up the circadian clock of cyanobacteria [4]. However, feedback loops of transcription are required to maintain the proper function of the oscillator [5]. More recently, the circadian oscillation was also found in the growth of Pseudomonas putida in the soil [6] and Bacillus subtilis [7], which is widely used as a probiotic additive in the animal production industry. Increasing studies reveal that the gut microbiota also exhibits robust diurnal rhythmicity at the compartmental, compositional, and functional levels in mice and humans [8][9][10][11][12]. As the primary microbial metabolites, the short-chain fatty acids (SCFAs) also undergo rhythmic oscillations [13][14][15]. However, the mechanism for the formation of microbial rhythmicity is not yet understood. Notably, the deletion of the host circadian clock gene resulted in the disrupted rhythmicity of the gut microbiota [10][16]. Whereas the disrupted gut microbiota rhythmicity was rescued by changing the feeding pattern. However, although playing a crucial role in maintaining the diurnal rhythmicity of gut microbiota, the host circadian clocks were not direct determinants of the gut microbial rhythmicity [10].
2. The Diurnal Rhythmicity of Gut Microbiota
In recent years, abundant evidence has indicated that the gut microbiota, its distribution, functions, and metabolites, underwent robust fluctuations [8][9][10][11][13][17]. The oral microbiome of humans also exhibited rhythmicity over a day [18]. More specifically, populations from different geographical areas have differential microbial fluctuation modes [12]. Further, similar to the compartmentalization character of microbial composition, the diurnal rhythmicity of the gut microbiota also exhibited inconsistencies between different regions of the intestine [13]. Unexpectedly, even in the same intestinal segment, the diurnal pattern of the luminal microorganisms differed from that of mucosal microbiota. Besides, the microscopic observations and quantitative PCR results have confirmed that more bacteria colonized the epithelial layer in the dark phase than in the light phase in mice [10]. It is worthy to note that different taxa dominated the gut microbial community at different times in the day. Firmicutes and Bacteroidetes were respectively prevalent at day and night in humans and mice [12][16]. As one of the most important commensal bacteria, the relative abundance of Lactobacillus was higher in the daytime than that in the nighttime [8]. In addition, over 20% of the microbial genes exhibited robust oscillations [8][10]. Regarding bacterial functions, pathways concerning DNA repair, cell growth, and energy metabolism were higher in the dark phase, while pathways related to detoxification, motility, and environmental sensing bloomed during the light phase [8]. Consistently, SCFAs also exhibited diurnal fluctuation, especially for acetate and butyrate [19].
3. The Influencers of the Gut Microbial Rhythmicity
Just like the gut microbiota configuration, the gut microbiota rhythmicity is affected by many factors such as nutritional factors (including nutrients level, antibiotics, feed additives, and diet composition), management factors (including feeding time and lighting regime), environmental factors, as well as host circadian rhythmicity and host physiology. The host circadian clock undoubtedly affects microbial rhythmicity. Related content will be discussed in the following section. Remarkably, high-fat, high-sucrose, and antibiotic-supplement diets have attenuated the rhythmic oscillation of the gut microbiota [9][10][20]. As a primary sulfur amino acid, methionine is an essential amino acid and has fundamental implications for maintaining energy-metabolism homeostasis. A methionine-restricted diet has potently alleviated inflammation in obese mice. Interestingly, the methionine-restricted diet has improved the disrupted rhythmicity of the gut microbiota induced by a high-fat diet [15]. Light is one of the most important determinants of host circadian rhythmicity. A reversing lighting regime has led to an antiphase oscillation of the most dominant microbe. Whereas a constantly dark regime has reduced the quantities of cyclical OTUs [21][22]. Further, Lu and Lee (2019) found that the light regime may entrain the rhythmicity of gut microbiota through intrinsically photosensitive retinal ganglion cells [23]. Also, supplementation of oolong tea extract can partially rescue the lost rhythmicity caused by the constantly dark regime [22]. Besides, gender is a possible factor affecting the rhythmicity of the intestinal microbiota. As one of the most dominant phyla, the diurnal rhythmicity of Bacteroidetes in relative abundance was more robust in female mice than in male mice [16]. Moreover, possible physiological and pathophysiological factors were also related to the gut microbiota rhythmicity. It is interesting to note that the rhythmicity of the oral microbiome was abolished when incubating the saliva in vitro, which implies the indispensable role of the host in maintaining the normal microbial rhythmicity [18]. In addition, individuals suffering from obstructive sleep apnea underwent an abnormal microbial oscillating pattern and metabolome [24]. Notably, compared with normal healthy people, people with obesity had damping rhythmicity of gut microbiota [12]. Sleeve gastrectomy is the most popular bariatric procedure worldwide. The dampened diurnal oscillation of gut microbiota in the obese mice induced by a high-fat diet was improved by the sleeve gastrectomy [25].
Interestingly, certain environmental factors would also affect the normal gut rhythmicity. For example, in a simulated space environment, the integrated low air pressure, noise, and weightlessness condition has dissimilatory impacts on the diurnal oscillation of gut microbiota [26]. The influencers of the gut microbiota rhythmicity are summarized in Figure 1.
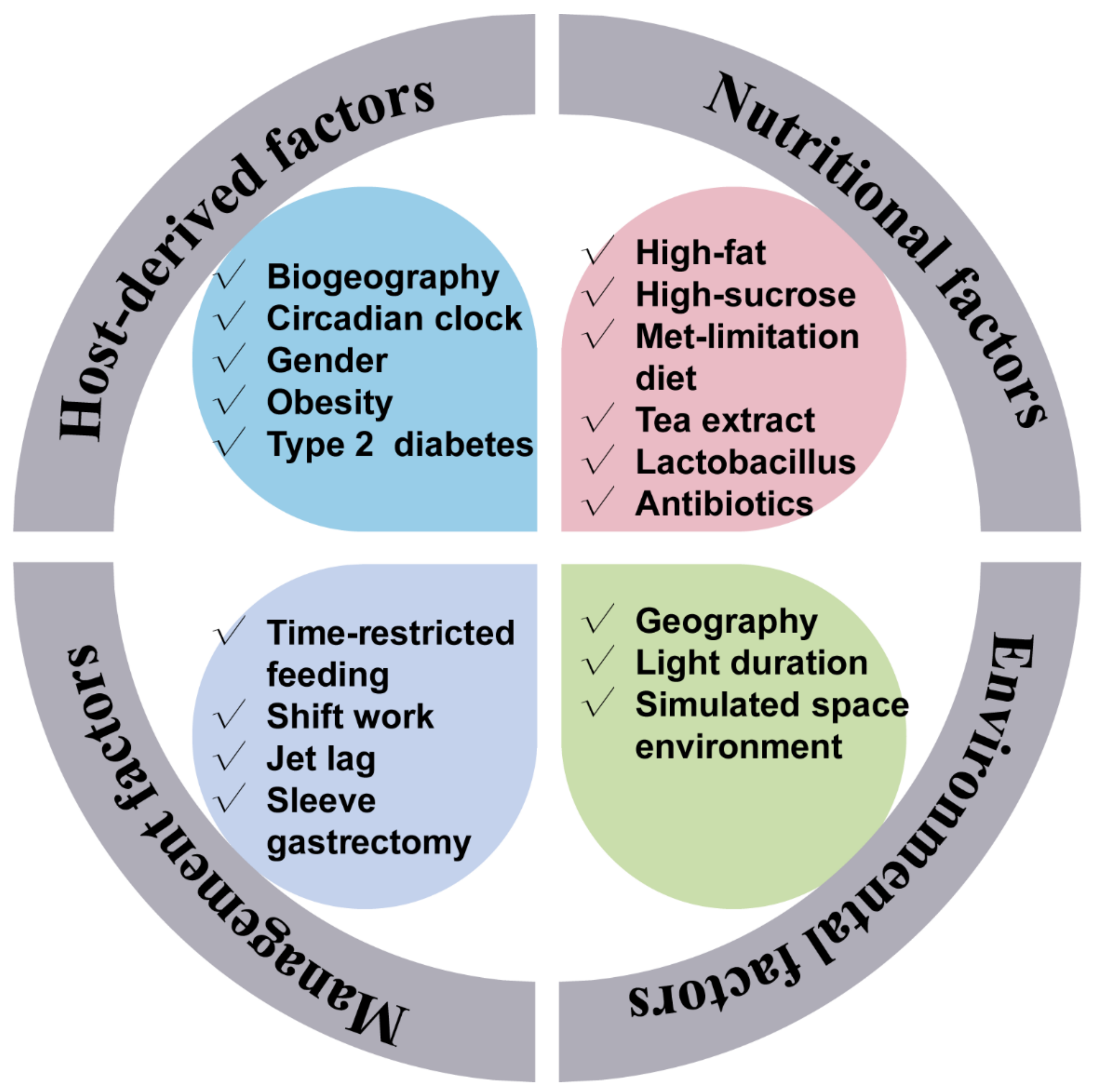 Figure 1. Factors that affect the gut microbiota rhythmicity. The gut microbiota rhythmicity was susceptible to many factors such as nutritional factors (including diet composition, nutrient levels, antibiotics, as well as feed additives), management factors (including feeding time, lighting regime), environmental factors, as well as host-derived factors.
Figure 1. Factors that affect the gut microbiota rhythmicity. The gut microbiota rhythmicity was susceptible to many factors such as nutritional factors (including diet composition, nutrient levels, antibiotics, as well as feed additives), management factors (including feeding time, lighting regime), environmental factors, as well as host-derived factors.4. The Gut Microbiota Rhythmicity Is Dynamically Intertwined with the Host Circadian Rhythm
Circadian rhythmicity is an important physiological phenomenon that plays an important role in maintaining metabolic homeostasis and sustaining normal life in prokaryotes and eukaryotes. The circadian clock in mammals consists of a master clock located in the suprachiasmatic nucleus of the hypothalamus and peripheral clocks located in other tissues of the body such as the liver and intestine [27]. The mammalian circadian clock is a self-sustaining, interlocked transcription-translation feedback loop of a network of genes including Bmal1, Clock, period circadian protein gene (Per1, Per2, and Per3), and cryptochrome gene (Cry1 and Cry2) [28]. Intriguingly, the microbiota in the intestine can co-evolve with the host and play an indispensable role in host physiology and metabolism. Importantly, intestinal dysbiosis and circadian rhythm disruption are associated with obesity, metabolic syndrome, and inflammatory bowel disease. The light signal is the main stimulus synchronizing the master clock with the external environment. Whereas the peripheral clocks can be entrained by other signals such as diet, feeding regime, hormones, and microbiota-derived SCFAs apart from the regulation of the master clock [29][30]. The disorganization of the core clock altered the microbial composition, which may further result in intestinal dysbiosis and inflammatory diseases in mice [31]. The mammal circadian clock orchestrates the behavioral and physiological processes throughout the day. Essentially, both the epigenetics and transcriptome showed a significant rhythmic fluctuation. More specifically, about 45% of the gene expressed in a diurnal manner. These dynamic expressions of genes further determined the rhythmic variation of most physiological processes [32][33]. Interdependence cross talks between the gut microbiota and the host circadian clock play a critical role in maintaining the normal circadian rhythmicity of the host and the gut microbial community (Figure 2).
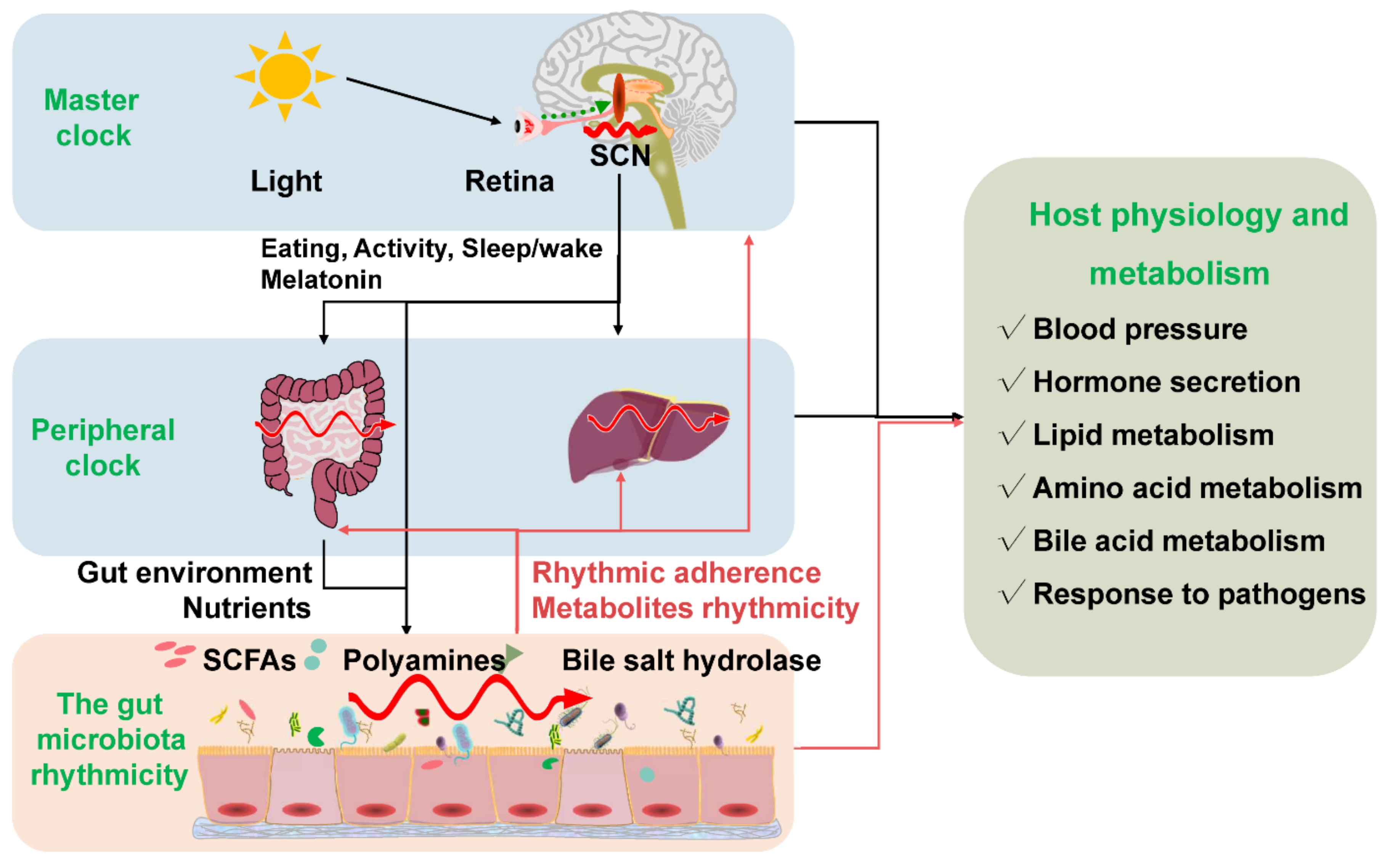 Figure 2. The gut microbiota rhythmicity is dynamically intertwined with the host’s circadian rhythm. The gut microbial rhythmicity works together with the host circadian system to maintain the normal rhythmicity of the host physiology and metabolism. Entrained by environmental factors (especially light cues), the master clock located in the SCN drives the host activities, such as eating behaviors, and sleep/wake rhythms to synchronize with the environment’s rhythmic changes. Through the microbiota–gut–brain axis and the gut–liver axis, the host master clock affects the rhythmicity of the gut microbiota and its metabolites (including SCFAs, polyamines, bile salt hydrolase). Interestingly, the gut microbiota and their metabolites could react to the normal transcriptomes and metabolism of the host circadian clock systems, including the master clock and the peripheral clock. SCN = suprachiasmatic nucleus; SCFAs = short-chain fatty acids.
Figure 2. The gut microbiota rhythmicity is dynamically intertwined with the host’s circadian rhythm. The gut microbial rhythmicity works together with the host circadian system to maintain the normal rhythmicity of the host physiology and metabolism. Entrained by environmental factors (especially light cues), the master clock located in the SCN drives the host activities, such as eating behaviors, and sleep/wake rhythms to synchronize with the environment’s rhythmic changes. Through the microbiota–gut–brain axis and the gut–liver axis, the host master clock affects the rhythmicity of the gut microbiota and its metabolites (including SCFAs, polyamines, bile salt hydrolase). Interestingly, the gut microbiota and their metabolites could react to the normal transcriptomes and metabolism of the host circadian clock systems, including the master clock and the peripheral clock. SCN = suprachiasmatic nucleus; SCFAs = short-chain fatty acids.The host circadian clock was one of the most important factors that can regulate the microbial rhythmicity. It should be noted that the depletion of the circadian clock gene (Bmal1 or Per1/2) has abrogated the rhythmic oscillations of the gut microbiota [8][16]. Moreover, the rhythmicity of microbial metabolites (e.g., SCFAs) was subsequently abolished in Bmal1 gene depletion mice [13][14]. Consistently, chronically disrupted circadian rhythm induced by shifting the light: dark regime, jet lag, and shift work have influenced the oscillation pattern of gut microbiota [8]. However, the lost rhythmicity of gut microbiota and its metabolites is rescued by manipulating the feeding patterns. The findings suggest that the diurnal rhythmicity of gut microbiota can independently exist without the circadian clock [10][13]. The possible explanation was that a disrupted circadian clock affected the diurnal feeding rhythm in mice [34], which further changed the composition and diurnal rhythmicity of the gut microbiota [8][9][16].
In turn, the rhythmicity of the gut microbiota can influence the normal function of the host circadian clock. For example, the rhythmic adherence of microbiota affects the circadian transcriptome in the intestine. The fluctuations of the microbial metabolites can rhythmically program the liver transcriptome [10]. Furthermore, using a mono-colonized bacteria, Thaiss et al. (2016) indicated that the rhythmic bacterial adherence, not the presence of gut microbiota, programmed the rhythmic transcription in the peripheral circadian clock [10]. Notably, the expression of the core clock genes was disrupted in germ-free mice with decreased Bmal1 and Cry1 transcripts and increased Per1 and Per2 transcripts [35].
SCFAs might be one of the possible synchronizers of peripheral circadian clocks [11]. Both in-vivo and in-vitro studies indicated that the administration of SCFAs could indirectly shift the oscillation phase of the peripheral clock in the liver in a rhythmic manner [19]. Thus, SCFAs might be essential in the dynamic interactions between gut microbiota and the circadian clock in maintaining the homeostasis of normal physiology and synchronizing circadian activities. Through histone deacetylase, the gut microbiota can rhythmically regulate the host metabolism and affect the diurnal oscillations of the host physiology and the susceptibility to pathologies [36]. The direct evidence was that the gene expression profile in the liver of the germ-free mice was different from that in the normal mice despite the diet [11]. Intriguingly, microbial butyrate can regulate the rhythm patterns of Per2 and Bmal1 in the peripheral circadian clocks, implying that the microbiota indirectly modulates circadian rhythms via metabolites [11]. In addition, gut microbiota can regulate the intrinsic Nfil3 circadian rhythms of the intestinal epithelial cell through type 3 innate lymphoid cells [37].
Polyamines are supposed to be primarily derived from diet and the gut microbiota, as the limitation of polyamines from the diet or microbiota has significantly decreased the circulating polyamine [38][39][40]. Compelling evidence suggests that polyamines, as pleiotropic signaling molecules, have been involved in various physiological and pathological processes [41][42]. More noteworthy, polyamines exhibit diurnal fluctuation. Changing the levels of polyamines could modulate the circadian period by regulating the interaction between the repressors of core clock PER2 and CRY1 both in in-vivo and in-vitro studies [17]. In addition to participating in carbohydrate metabolism and amino acid metabolism, the gut microbiota also plays an essential role in lipid metabolism, remarkably so, in pathways concerning the synthesis and oxidation of fatty acids [43][44]. The lipid concentrations in the liver and bloodstream were also oscillating rhythmically [45][46]. Choline trimethylamine-lyase derived from the gut microbiota is a rate-limiting enzyme transforming choline into trimethylamine. Surprisingly, inhibiting choline trimethylamine-lyase significantly reduced atherosclerosis and thrombosis and improved obesity in mice through remodeling the host peripheral circadian clocks [47][48][49].
Moreover, as emulsifiers in lipid metabolism, bile acids play critical roles in maintaining host metabolic homeostasis due to regulating the metabolism pathways related to cholesterol, triglyceride, and glucose metabolism [50][51]. Bile metabolism is in a time-of-day manner due to the need to coordinate metabolic responses to food intake, enabling the esterification and absorption of dietary fats and lipids [52][53]. The metabolism of bile acids, primarily secondary bile acids, is regulated by the gut microbiota [54][55]. Also, relevant findings suggested that the serum bile acids peaked at the beginning and end of the dark phase, while the serum secondary bile acids peaked at the beginning of the dark phase [56][57]. In addition, the microbial bile salt hydrolase significantly regulated the transcription of circadian rhythm genes such as Dbp and Per1/2 in the liver or small intestine [58].
This entry is adapted from the peer-reviewed paper 10.3390/ani12131677
References
- Green, C.B.; Takahashi, J.S.; Bass, J. The meter of metabolism. Cell 2008, 134, 728–742.
- Mitsui, A.; Kumazawa, S.; Takahashi, A.; Ikemoto, H.; Cao, S.; Arai, T. Strategy by which nitrogen-fixing unicellular cyanobacteria grow photoautotrophically. Nature 1986, 323, 720–722.
- Stal, L.; Krumbein, W. Temporal separation of nitrogen fixation and photosynthesis in the filamentous, non-heterocystous cyanobacterium Oscillatoria sp. Arch. Microbiol. 1987, 149, 76–80.
- O’Neill, J.S.; Van Ooijen, G.; Dixon, L.E.; Troein, C.; Corellou, F.; Bouget, F.-Y.; Reddy, A.B.; Millar, A.J. Circadian rhythms persist without transcription in a eukaryote. Nature 2011, 469, 554–558.
- Teng, S.-W.; Mukherji, S.; Moffitt, J.R.; De Buyl, S.; O’shea, E.K. Robust circadian oscillations in growing cyanobacteria require transcriptional feedback. Science 2013, 340, 737–740.
- Soriano, M.I.; Roibás, B.; García, A.B.; Espinosa-Urgel, M. Evidence of circadian rhythms in non-photosynthetic bacteria? J. Circadian Rhythm. 2010, 8, 8.
- Sartor, F.; Eelderink-Chen, Z.; Aronson, B.; Bosman, J.; Hibbert, L.E.; Dodd, A.N.; Kovács, Á.T.; Merrow, M. Are there circadian clocks in non-photosynthetic bacteria? Biology 2019, 8, 41.
- Thaiss, C.A.; Zeevi, D.; Levy, M.; Zilberman-Schapira, G.; Suez, J.; Tengeler, A.C.; Abramson, L.; Katz, M.N.; Korem, T.; Zmora, N. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell 2014, 159, 514–529.
- Zarrinpar, A.; Chaix, A.; Yooseph, S.; Panda, S. Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metab. 2014, 20, 1006–1017.
- Thaiss, C.A.; Levy, M.; Korem, T.; Dohnalová, L.; Shapiro, H.; Jaitin, D.A.; David, E.; Winter, D.R.; Gury-BenAri, M.; Tatirovsky, E. Microbiota diurnal rhythmicity programs host transcriptome oscillations. Cell 2016, 167, 1495–1510.e12.
- Leone, V.; Gibbons, S.M.; Martinez, K.; Hutchison, A.L.; Huang, E.Y.; Cham, C.M.; Pierre, J.F.; Heneghan, A.F.; Nadimpalli, A.; Hubert, N. Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host Microbe 2015, 17, 681–689.
- Reitmeier, S.; Kiessling, S.; Clavel, T.; List, M.; Almeida, E.L.; Ghosh, T.S.; Neuhaus, K.; Grallert, H.; Linseisen, J.; Skurk, T. Arrhythmic gut microbiome signatures predict risk of type 2 diabetes. Cell Host Microbe 2020, 28, 258–272.e6.
- Segers, A.; Desmet, L.; Thijs, T.; Verbeke, K.; Tack, J.; Depoortere, I. The circadian clock regulates the diurnal levels of microbial short-chain fatty acids and their rhythmic effects on colon contractility in mice. Acta Physiol. 2019, 225, e13193.
- Segers, A.; Desmet, L.; Sun, S.; Verbeke, K.; Tack, J.; Depoortere, I. Night-time feeding of Bmal1−/− mice restores SCFA rhythms and their effect on ghrelin. J. Endocrinol. 2020, 245, 155–164.
- Wang, L.; Ren, B.; Hui, Y.; Chu, C.; Zhao, Z.; Zhang, Y.; Zhao, B.; Shi, R.; Ren, J.; Dai, X. Methionine Restriction Regulates Cognitive Function in High-Fat Diet-Fed Mice: Roles of Diurnal Rhythms of SCFAs Producing-and Inflammation-Related Microbes. Mol. Nutr. Food Res. 2020, 64, 2000190.
- Liang, X.; Bushman, F.D.; FitzGerald, G.A. Rhythmicity of the intestinal microbiota is regulated by gender and the host circadian clock. Proc. Natl. Acad. Sci. USA 2015, 112, 10479–10484.
- Zwighaft, Z.; Aviram, R.; Shalev, M.; Rousso-Noori, L.; Kraut-Cohen, J.; Golik, M.; Brandis, A.; Reinke, H.; Aharoni, A.; Kahana, C. Circadian clock control by polyamine levels through a mechanism that declines with age. Cell Metab. 2015, 22, 874–885.
- Takayasu, L.; Suda, W.; Takanashi, K.; Iioka, E.; Kurokawa, R.; Shindo, C.; Hattori, Y.; Yamashita, N.; Nishijima, S.; Oshima, K. Circadian oscillations of microbial and functional composition in the human salivary microbiome. DNA Res. 2017, 24, 261–270.
- Tahara, Y.; Yamazaki, M.; Sukigara, H.; Motohashi, H.; Sasaki, H.; Miyakawa, H.; Haraguchi, A.; Ikeda, Y.; Fukuda, S.; Shibata, S. Gut microbiota-derived short chain fatty acids induce circadian clock entrainment in mouse peripheral tissue. Sci. Rep. 2018, 8, 1395.
- Sun, S.; Araki, Y.; Hanzawa, F.; Umeki, M.; Kojima, T.; Nishimura, N.; Ikeda, S.; Mochizuki, S.; Oda, H. High sucrose diet-induced dysbiosis of gut microbiota promotes fatty liver and hyperlipidemia in rats. J. Nutr. Biochem. 2021, 93, 108621.
- Wu, G.; Tang, W.; He, Y.; Hu, J.; Gong, S.; He, Z.; Wei, G.; Lv, L.; Jiang, Y.; Zhou, H. Light exposure influences the diurnal oscillation of gut microbiota in mice. Biochem. Biophys. Res. Commun. 2018, 501, 16–23.
- Guo, T.; Ho, C.-T.; Zhang, X.; Cao, J.; Wang, H.; Shao, X.; Pan, D.; Wu, Z. Oolong tea polyphenols ameliorate circadian rhythm of intestinal microbiome and liver clock genes in mouse model. J. Agric. Food Chem. 2019, 67, 11969–11976.
- Lu, T.-H.; Lee, C.-C. Light regulates gut microbiome composition and rhythmicity through ipRGCs. Investig. Ophthalmol. Vis. Sci. 2019, 60, 5259.
- Allaband, C.; Lingaraju, A.; Martino, C.; Russell, B.; Tripathi, A.; Poulsen, O.; Dantas Machado, A.C.; Zhou, D.; Xue, J.; Elijah, E. Intermittent Hypoxia and Hypercapnia Alter Diurnal Rhythms of Luminal Gut Microbiome and Metabolome. Msystems 2021, 6, e00116–e00121.
- Shao, Y.; Shen, Q.; Hua, R.; Evers, S.S.; He, K.; Yao, Q. Effects of sleeve gastrectomy on the composition and diurnal oscillation of gut microbiota related to the metabolic improvements. Surg. Obes. Relat. Dis. 2018, 14, 731–739.
- Ma, H.; Gan, X.; Zhao, J.; Zhang, Y.; Li, S.; Kan, G.; Wang, B.; Zhang, P.; Ma, X.; Tian, H. Simulated Space Environmental Factors of Weightlessness, Noise and Low Air Pressure Differentially Affect the Circadian Rhythm and Gut Microbiome. 2021. Available online: https://doi.org/10.21203/rs.3.rs-362076/v1 (accessed on 12 January 2022).
- Moore, R.Y.; Eichler, V.B. Loss of a circadian adrenal corticosterone rhythm following suprachiasmatic lesions in the rat. Brain Res. 1972, 42, 201–206.
- Reppert, S.M.; Weaver, D.R. Molecular analysis of mammalian circadian rhythms. Annu. Rev. Physiol. 2001, 63, 647–676.
- Laermans, J.; Broers, C.; Beckers, K.; Vancleef, L.; Steensels, S.; Thijs, T.; Tack, J.; Depoortere, I. Shifting the circadian rhythm of feeding in mice induces gastrointestinal, metabolic and immune alterations which are influenced by ghrelin and the core clock gene Bmal1. PLoS ONE 2014, 9, e110176.
- Landgraf, D.; Tsang, A.H.; Leliavski, A.; Koch, C.E.; Barclay, J.L.; Drucker, D.J.; Oster, H. Oxyntomodulin regulates resetting of the liver circadian clock by food. eLife 2015, 4, e06253.
- Voigt, R.M.; Forsyth, C.B.; Green, S.J.; Mutlu, E.; Engen, P.; Vitaterna, M.H.; Turek, F.W.; Keshavarzian, A. Circadian disorganization alters intestinal microbiota. PLoS ONE 2014, 9, e97500.
- Badia, P.; Myers, B.; Boecker, M.; Culpepper, J.; Harsh, J. Bright light effects on body temperature, alertness, EEG and behavior. Physiol. Behav. 1991, 50, 583–588.
- Vandewalle, G.; Maquet, P.; Dijk, D.-J. Light as a modulator of cognitive brain function. Trends Cogn. Sci. 2009, 13, 429–438.
- Turek, F.W.; Joshu, C.; Kohsaka, A.; Lin, E.; Ivanova, G.; McDearmon, E.; Laposky, A.; Losee-Olson, S.; Easton, A.; Jensen, D.R. Obesity and metabolic syndrome in circadian Clock mutant mice. Science 2005, 308, 1043–1045.
- Mukherji, A.; Kobiita, A.; Ye, T.; Chambon, P. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell 2013, 153, 812–827.
- Kuang, Z.; Wang, Y.; Li, Y.; Ye, C.; Ruhn, K.A.; Behrendt, C.L.; Olson, E.N.; Hooper, L.V. The intestinal microbiota programs diurnal rhythms in host metabolism through histone deacetylase 3. Science 2019, 365, 1428–1434.
- Wang, Y.; Kuang, Z.; Yu, X.; Ruhn, K.A.; Kubo, M.; Hooper, L.V. The intestinal microbiota regulates body composition through NFIL3 and the circadian clock. Science 2017, 357, 912–916.
- Lotz, M.; König, T.; Ménard, S.; Gütle, D.; Bogdan, C.; Hornef, M.W. Cytokine-mediated control of lipopolysaccharide-induced activation of small intestinal epithelial cells. Immunology 2007, 122, 306–315.
- Morris, S.M., Jr. Recent advances in arginine metabolism: Roles and regulation of the arginases. Br. J. Pharmacol. 2009, 157, 922–930.
- Hibasami, H.; Hoffman, J.; Pegg, A. Decarboxylated S-adenosylmethionine in mammalian cells. J. Biol. Chem. 1980, 255, 6675–6678.
- Fan, P.; Song, P.; Li, L.; Huang, C.; Chen, J.; Yang, W.; Qiao, S.; Wu, G.; Zhang, G.; Ma, X. Roles of biogenic amines in intestinal signaling. Curr. Protein Pept. Sci. 2017, 18, 532–540.
- Sudo, N. Biogenic amines: Signals between commensal microbiota and gut physiology. Front. Endocrinol. 2019, 10, 504.
- Huang, S.-M.; Wu, Z.-H.; Li, T.-T.; Liu, C.; Han, D.-D.; Tao, S.-Y.; Pi, Y.; Li, N.; Wang, J.-J. Perturbation of the lipid metabolism and intestinal inflammation in growing pigs with low birth weight is associated with the alterations of gut microbiota. Sci. Total Environ. 2020, 719, 137382.
- Wu, Y.; Ma, N.; Song, P.; He, T.; Levesque, C.; Bai, Y.; Zhang, A.; Ma, X. Grape seed proanthocyanidin affects lipid metabolism via changing gut microflora and enhancing propionate production in weaned pigs. J. Nutr. 2019, 149, 1523–1532.
- Neufeld-Cohen, A.; Robles, M.S.; Aviram, R.; Manella, G.; Adamovich, Y.; Ladeuix, B.; Nir, D.; Rousso-Noori, L.; Kuperman, Y.; Golik, M. Circadian control of oscillations in mitochondrial rate-limiting enzymes and nutrient utilization by PERIOD proteins. Proc. Natl. Acad. Sci. USA 2016, 113, E1673–E1682.
- Adamovich, Y.; Rousso-Noori, L.; Zwighaft, Z.; Neufeld-Cohen, A.; Golik, M.; Kraut-Cohen, J.; Wang, M.; Han, X.; Asher, G. Circadian clocks and feeding time regulate the oscillations and levels of hepatic triglycerides. Cell Metab. 2014, 19, 319–330.
- Roberts, A.B.; Gu, X.; Buffa, J.A.; Hurd, A.G.; Wang, Z.; Zhu, W.; Gupta, N.; Skye, S.M.; Cody, D.B.; Levison, B.S. Development of a gut microbe–targeted nonlethal therapeutic to inhibit thrombosis potential. Nat. Med. 2018, 24, 1407–1417.
- Organ, C.L.; Li, Z.; Sharp III, T.E.; Polhemus, D.J.; Gupta, N.; Goodchild, T.T.; Tang, W.W.; Hazen, S.L.; Lefer, D.J. Nonlethal Inhibition of Gut Microbial Trimethylamine N-oxide Production Improves Cardiac Function and Remodeling in a Murine Model of Heart Failure. J. Am. Heart Assoc. 2020, 9, e016223.
- Schugar, R.C.; Gliniak, C.M.; Osborn, L.J.; Massey, W.; Sangwan, N.; Horak, A.; Banerjee, R.; Orabi, D.; Helsley, R.N.; Brown, A.L. Gut microbe-targeted choline trimethylamine lyase inhibition improves obesity via rewiring of host circadian rhythms. eLife 2022, 11, e63998.
- Ge, X.; Wang, A.; Ying, Z.; Zhang, L.; Su, W.; Cheng, K.; Feng, C.; Zhou, Y.; Zhang, L.; Wang, T. Effects of diets with different energy and bile acids levels on growth performance and lipid metabolism in broilers. Poult. Sci. 2019, 98, 887–895.
- Duparc, T.; Plovier, H.; Marrachelli, V.G.; Van Hul, M.; Essaghir, A.; Ståhlman, M.; Matamoros, S.; Geurts, L.; Pardo-Tendero, M.M.; Druart, C. Hepatocyte MyD88 affects bile acids, gut microbiota and metabolome contributing to regulate glucose and lipid metabolism. Gut 2017, 66, 620–632.
- Harris, V.; Ali, A.; Fuentes, S.; Korpela, K.; Kazi, M.; Tate, J.; Parashar, U.; Wiersinga, W.J.; Giaquinto, C.; de Weerth, C. Rotavirus vaccine response correlates with the infant gut microbiota composition in Pakistan. Gut Microbes 2018, 9, 93–101.
- Harris, V.C.; Haak, B.W.; Handley, S.A.; Jiang, B.; Velasquez, D.E.; Hykes, B.L., Jr.; Droit, L.; Berbers, G.A.; Kemper, E.M.; van Leeuwen, E.M. Effect of antibiotic-mediated microbiome modulation on rotavirus vaccine immunogenicity: A human, randomized-control proof-of-concept trial. Cell Host Microbe 2018, 24, 197–207.e194.
- Chen, M.L.; Takeda, K.; Sundrud, M.S. Emerging roles of bile acids in mucosal immunity and inflammation. Mucosal Immunol. 2019, 12, 851–861.
- Theriot, C.M.; Bowman, A.A.; Young, V.B. Antibiotic-induced alterations of the gut microbiota alter secondary bile acid production and allow for Clostridium difficile spore germination and outgrowth in the large intestine. MSphere 2016, 1, e00045-15.
- Zhang, Y.-K.J.; Guo, G.L.; Klaassen, C.D. Diurnal variations of mouse plasma and hepatic bile acid concentrations as well as expression of biosynthetic enzymes and transporters. PLoS ONE 2011, 6, e16683.
- Eggink, H.M.; Oosterman, J.E.; de Goede, P.; de Vries, E.M.; Foppen, E.; Koehorst, M.; Groen, A.K.; Boelen, A.; Romijn, J.A.; la Fleur, S.E. Complex interaction between circadian rhythm and diet on bile acid homeostasis in male rats. Chronobiol. Int. 2017, 34, 1339–1353.
- Joyce, S.A.; MacSharry, J.; Casey, P.G.; Kinsella, M.; Murphy, E.F.; Shanahan, F.; Hill, C.; Gahan, C.G. Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc. Natl. Acad. Sci. USA 2014, 111, 7421–7426.
This entry is offline, you can click here to edit this entry!
