Current influenza vaccine candidates, for potential use in vaccine manufacturing, are reassortants of master donor virus (MDV) with wild-type (WT) virus that is antigenically similar to the recommended strain. MDVs have all the necessary characteristics for the type of vaccines of which they are intended. Two types of MDVs are used in the preparation of influenza vaccines—high-yielding donors for IIV and temperature-sensitive (ts) and cold-adapted (ca) donors of attenuation—for LAIV. There are a number of main features of WT influenza virus that may dramatically affect different aspects of the preparation of egg-derived live attenuated vaccine candidates and their effectiveness.
- influenza vaccine
- classical reassortment
- reassortant
1. Naturally Occurring Temperature-Sensitive WT Influenza Viruses
2. Naturally Occurring Cold-Adapted WT Influenza Viruses
3. Sensitivity of WT Viruses to Nonspecific Thermostable Serum γ-Inhibitors
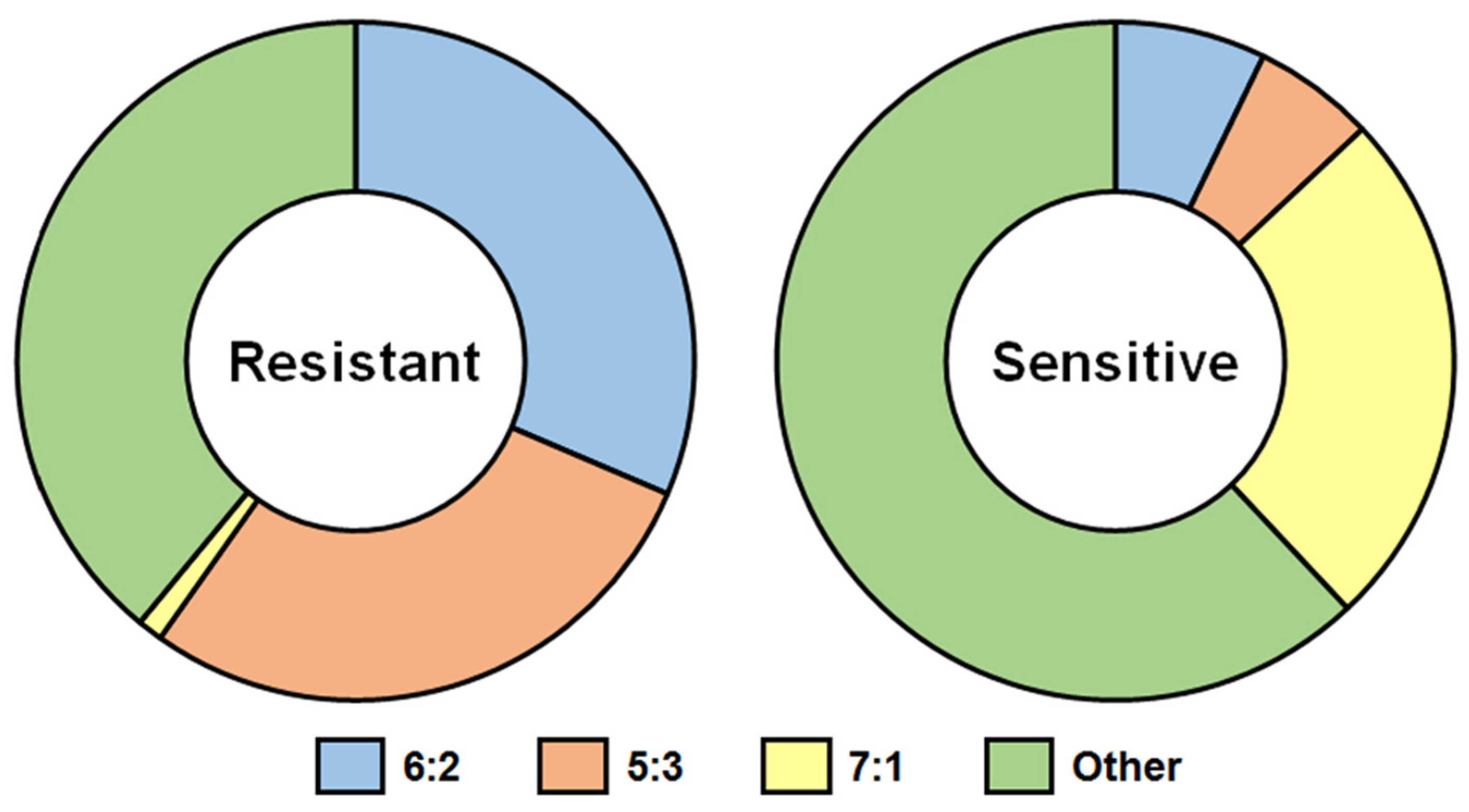
4. Infectivity of WT Viruses and LAIV Candidates
This entry is adapted from the peer-reviewed paper 10.3390/ijms23126815
References
- Mills, J.; Chanock, V.; Chanock, R.M. Temperature-sensitive mutants of influenza virus. I. Behavior in tissue culture and in experimental animals. J. Infect. Dis. 1971, 123, 145–157.
- Maassab, H.F.; DeBorde, D.C. Development and characterization of cold-adapted viruses for use as live virus vaccines. Vaccine 1985, 3, 355–369.
- Oxford, J.S.; Corcoran, T.; Schild, G.C. Naturally occurring temperature-sensitive influenza A viruses of the H1N1 and H3N2 subtypes. J. Gen. Virol. 1980, 48, 383–389.
- Chu, C.M.; Tian, S.F.; Ren, G.F.; Zhang, Y.M.; Zhang, L.X.; Liu, G.Q. Occurrence of temperature-sensitive influenza A viruses in nature. J. Virol. 1982, 41, 353–359.
- Polezhaev, F.I.; Aleksandrova, G.I. Isolation of temperature-sensitive strains of the influenza virus in the epidemic caused by the A/Victoria virus in 1975–1976. Vopr. Virusol. 1979, 4, 430.
- Zhang, Y.M.; Tian, S.F.; Zhu, J.M. Identification of naturally occurring temperature-sensitive strains of influenza A virus and location of their genetic lesions. Sci. Sin. B 1982, 25, 411–419.
- Richman, D.D.; Murphy, B.R. The association of the temperature-sensitive phenotype with viral attenuation in animals and humans: Implications for the development and use of live virus vaccines. Rev. Infect. Dis. 1979, 1, 413–433.
- Kiseleva, I.; Larionova, N. (Eds.) Influenza virus ecology and evolution. In Influenza: A Century of Research; Bentham Science Publisher: Sharjah, United Arab Emirates, 2021; pp. 63–97.
- Larionova, N.V.; Kiseleva, I.V.; Rudenko, L.G. Evolution of influenza viruses based on sensitivity to temperature of replication. J. Microbiol. Epidemiol. Immunobiol. 2019, 6, 47–55.
- Rudenko, L.G.; Kiseleva, I.V.; Larionova, N.V.; Grigorieva, E.P.; Naikhin, A.N. Analysis of some factors influencing immunogenicity of live cold–adapted reassortant influenza vaccines. In Proceedings of the Options for the Control of Influenza V, Okinawa, Japan, 6–9 October 2003; pp. 542–546.
- Kiseleva, I.V.; Voeten, J.T.; Teley, L.C.; Larionova, N.V.; Drieszen-van der Cruijsen, S.K.; Basten, S.M.; Heldens, J.G.; van den Bosch, H.; Rudenko, L.G. PB2 and PA genes control the expression of the temperature-sensitive phenotype of cold-adapted B/USSR/60/69 influenza master donor virus. J. Gen. Virol. 2010, 91, 931–937.
- Rogers, G.N.; D’Souza, B.L. Receptor binding properties of human and animal H1 influenza virus isolates. Virology 1989, 173, 317–322.
- Rogers, G.N.; Pritchett, T.J.; Lane, J.L.; Paulson, J.C. Differential sensitivity of human, avian, and equine influenza A viruses to a glycoprotein inhibitor of infection: Selection of receptor specific variants. Virology 1983, 131, 394–408.
- Kiseleva, I.; Larionova, N.; Fedorova, E.; Bazhenova, E.; Dubrovina, I.; Isakova-Sivak, I.; Rudenko, L. Contribution of neuraminidase of influenza viruses to the sensitivity to sera inhibitors and reassortment efficiency. Open Microbiol. J. 2014, 8, 59–70.
- Yang, J.R.; Huang, Y.P.; Chang, F.Y.; Hsu, L.C.; Lin, Y.C.; Huang, H.Y.; Wu, F.T.; Wu, H.S.; Liu, M.T. Phylogenetic and evolutionary history of influenza B viruses, which caused a large epidemic in 2011–2012, Taiwan. PLoS ONE 2012, 7, e47179.
- Rota, P.A.; Wallis, T.R.; Harmon, M.W.; Rota, J.S.; Kendal, A.P.; Nerome, K. Cocirculation of two distinct evolutionary lineages of influenza type B virus since 1983. Virology 1990, 175, 59–68.
- Larionova, N.; Kiseleva, I.; Isakova, I.; Litvinova, O.; Rudenko, L. Naturally occuring temperature-sensitive strains of influenza B virus. Vopr. Virusol. 2006, 51, 38–41.
- Shcherbik, S.; Pearce, N.; Kiseleva, I.; Larionova, N.; Rudenko, L.; Xu, X.; Wentworth, D.E.; Bousse, T. Implementation of new approaches for generating conventional reassortants for live attenuated influenza vaccine based on Russian master donor viruses. J. Virol. Methods 2016, 227, 33–39.
- Wareing, M.D.; Marsh, G.A.; Tannock, G.A. Preparation and characterisation of attenuated cold-adapted influenza A reassortants derived from the A/Leningrad/134/17/57 donor strain. Vaccine 2002, 20, 2082–2090.
- Shcherbik, S.V.; Pearce, N.C.; Levine, M.L.; Klimov, A.I.; Villanueva, J.M.; Bousse, T.L. Rapid strategy for screening by pyrosequencing of influenza virus reassortants-candidates for live attenuated vaccines. PLoS ONE 2014, 9, e92580.
- Fulvini, A.A.; Ramanunninair, M.; Le, J.; Pokorny, B.A.; Arroyo, J.M.; Silverman, J.; Devis, R.; Bucher, D. Gene constellation of influenza A virus reassortants with high growth phenotype prepared as seed candidates for vaccine production. PLoS ONE 2011, 6, e20823.
- Trifkovic, S.; Gilbertson, B.; Fairmaid, E.; Cobbin, J.; Rockman, S.; Brown, L.E. Gene segment interactions can drive the emergence of dominant yet suboptimal gene constellations during influenza virus reassortment. Front. Microbiol. 2021, 12, 683152.
- Kiseleva, I.V.; Larionova, N.V.; Fedorova, E.A.; Isakova-Sivak, I.N.; Rudenko, L.G. New methodological approaches in the development of Russian live attenuated vaccine for pandemic influenza. Transl. Biomed. 2015, 6, 1–9.
- Larionova, N.; Kiseleva, I.; Isakova-Sivak, I.; Rekstin, A.; Dubrovina, I.; Bazhenova, E.; Ross, T.M.; Swayne, D.; Gubareva, L.; Tsvetnitsky, V.; et al. Live attenuated influenza vaccines against highly pathogenic H5N1 avian influenza: Development and preclinical characterization. J. Vaccines Vaccin. 2013, 4, 208.
