Exosomes are membranous structures secreted by nearly all cell types. As critical messengers for intercellular communication, exosomes deliver bioactive cargoes to recipient cells and are involved in multiple physiopathological processes, including immunoregulation. This pioneering research revealed that cancer cells release programmed death-ligand 1-positive exosomes into the circulation to counter antitumor immunity systemically via T cells. Tumor cell-derived exosomes (TDEs) also play an immunosuppressive role in other immunocytes, including dendritic cells (DCs), macrophages, natural killer (NK) cells, and myeloid-derived suppressor cells (MDSCs). Moreover, exosomes secreted by nontumor cells in the tumor microenvironments (TMEs) also exert immunosuppressive effects.
- exosomes
- tumor microenvironments
- immunosuppression
- tumor cell
- immune cell
1. Introduction
2. Life Course of Exosomes
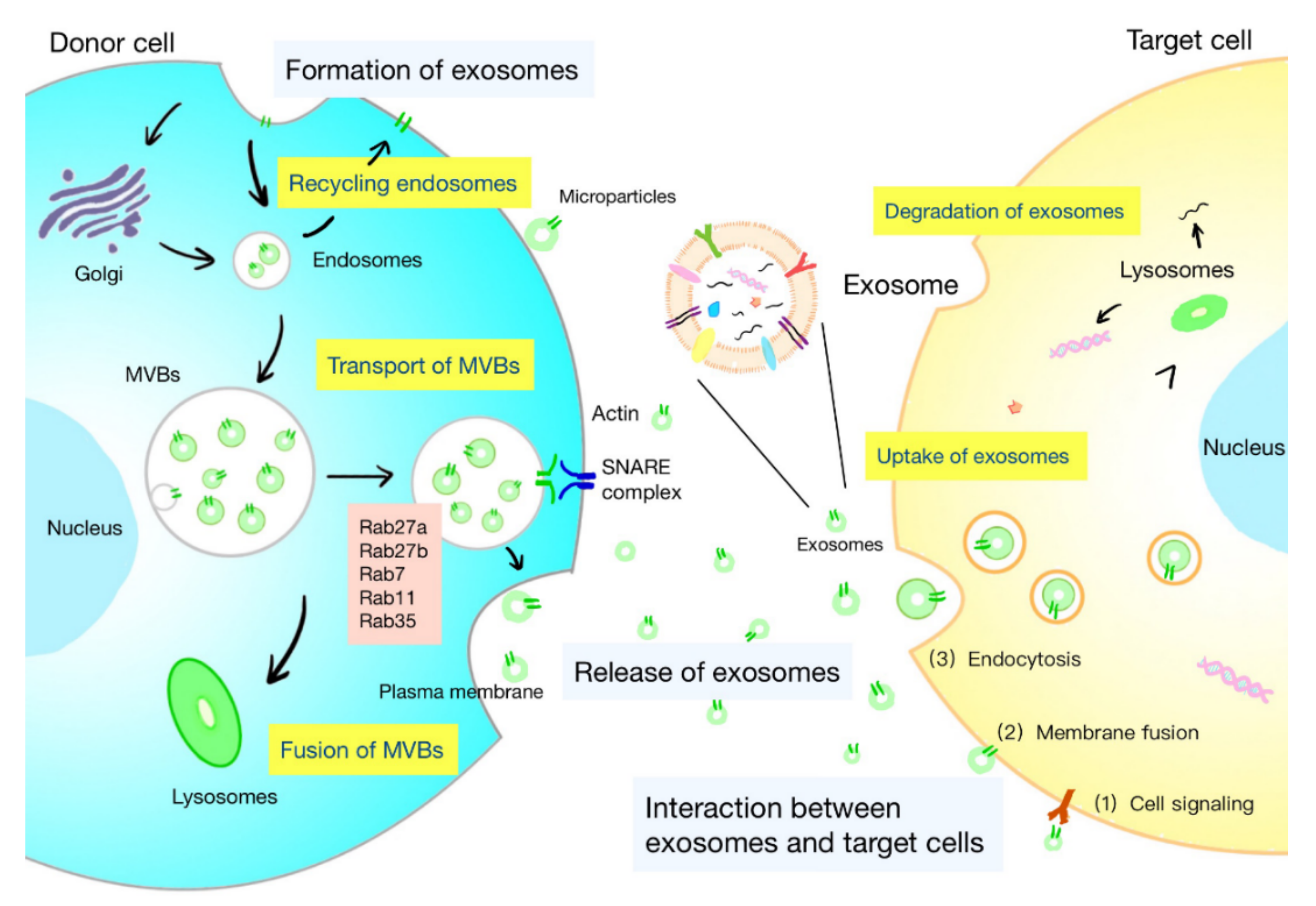
2.1. Formation of Exosomes
2.2. Release of Exosomes
2.3. Interaction between Exosomes and Target Cells
3. Exosomes as Cancer Biomarkers
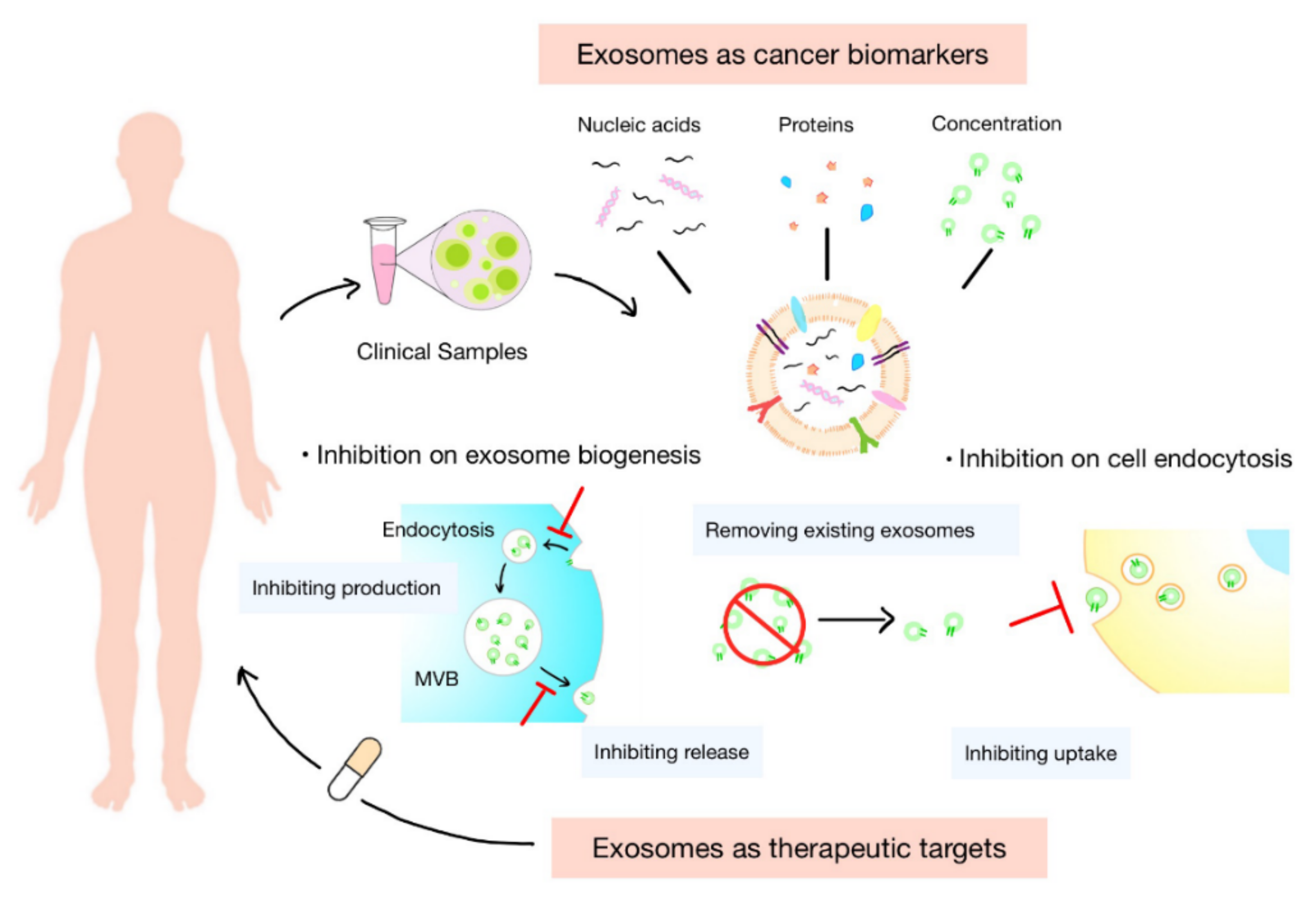
3.1. Concentration of Exosomes
3.2. Nucleic Acids in Exosomes
3.3. Proteins in Exosomes
3.4. Isolation and Identification of Exosomes
4. Exosomes as Therapeutic Targets
4.1. Inhibition of Biogenesis of Exosomes
4.2. Inhibition of Endocytosis of Exosomes by Recipient Cells
This entry is adapted from the peer-reviewed paper 10.3390/cells11121946
References
- Van Niel, G.; D’Angelo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228.
- Théry, C.; Witwer, W.K. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750.
- Sung, B.H.; Parent, C.A. Extracellular vesicles: Critical players during cell migration. Dev. Cell 2021, 56, 1861–1874.
- Mashouri, L.; Yousefi, H. Exosomes: Composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol. Cancer 2019, 18, 75.
- Yang, E.; Wang, X. Exosome-mediated metabolic reprogramming: The emerging role in tumor microenvironment remodeling and its influence on cancer progression. Signal Transduct. Target. Ther. 2020, 5, 242.
- Kalluri, R. The biology and function of exosomes in cancer. J. Clin. Investig. 2016, 126, 1208–1215.
- Zhou, X.; Xie, F. The function and clinical application of extracellular vesicles in innate immune regulation. Cell. Mol. Immunol. 2020, 17, 323–334.
- Alonso, R.; Rodríguez, M.C. Diacylglycerol kinase alpha regulates the secretion of lethal exosomes bearing Fas ligand during activation-induced cell death of T lymphocytes. J. Biol. Chem. 2005, 280, 28439–28450.
- Laulagnier, K.; Grand, D. PLD2 is enriched on exosomes and its activity is correlated to the release of exosomes. FEBS Lett. 2004, 572, 11–14.
- D’Souza-Schorey, C.; Schorey, J.S. Regulation and mechanisms of extracellular vesicle biogenesis and secretion. Essays Biochem. 2018, 62, 125–133.
- Mathieu, M.; Martin-Jaular, L. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019, 21, 9–17.
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383.
- Jia, Y.; Chen, Y. Exosome: Emerging biomarker in breast cancer. Oncotarget. 2017, 8, 41717–41733.
- Melo, S.A.; Luecke, L.B. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015, 523, 177–182.
- Kok, V.C.; Yu, C.C. Cancer-derived exosomes: Their role in cancer biology and biomarker development. Int. J. Nanomed. 2020, 15, 8019–8036.
- Lunavat, T.R.; Cheng, L. Small RNA deep sequencing discriminates subsets of extracellular vesicles released by melanoma cells--Evidence of unique microRNA cargos. RNA Biol. 2015, 12, 810–823.
- Whiteside, T.L. The potential of tumor-derived exosomes for noninvasive cancer monitoring. Expert Rev. Mol. Diagn. 2015, 15, 1293–1310.
- Wu, Y.; Zeng, H. A circulating exosome RNA signature is a potential diagnostic marker for pancreatic cancer, a systematic study. Cancers 2021, 13, 2565.
- Xu, H.; Dong, X. Serum exosomal hnRNPH1 mRNA as a novel marker for hepatocellular carcinoma. Clin. Chem. Lab. Med. 2018, 56, 479–484.
- Yokota, Y.; Noda, T. Serum exosomal miR-638 is a prognostic marker of HCC via downregulation of VE-cadherin and ZO-1 of endothelial cells. Cancer Sci. 2021, 112, 1275–1288.
- Yugawa, K.; Yoshizumi, T. Cancer-associated fibroblasts promote hepatocellular carcinoma progression through downregulation of exosomal miR-150-3p. Eur. J. Surg. Oncol. 2021, 47, 384–393.
- Shi, M.; Jiang, Y. Decreased levels of serum exosomal miR-638 predict poor prognosis in hepatocellular carcinoma. J. Cell. Biochem. 2018, 119, 4711–4716.
- Wang, T.; Zhu, H. Serum exosomal long noncoding RNA CRNDE as a prognostic biomarker for hepatocellular carcinoma. J. Clin. Lab. Anal. 2021, 35, e23959.
- Yu, S.; Li, Y. Plasma extracellular vesicle long RNA profiling identifies a diagnostic signature for the detection of pancreatic ductal adenocarcinoma. Gut 2020, 69, 540–550.
- Huang, T.; Deng, C.X. Current progresses of exosomes as cancer diagnostic and prognostic biomarkers. Int. J. Biol. Sci. 2019, 15, 1–11.
- Beccard, I.J.; Hofmann, L. Immune suppressive effects of plasma-derived exosome populations in head and neck cancer. Cancers 2020, 12, 7.
- Sharma, P.; Diergaarde, B. Melanoma cell-derived exosomes in plasma of melanoma patients suppress functions of immune effector cells. Sci. Rep. 2020, 10, 92.
- Wei, Q.; Feng, H. Serum exosomal EphA2 is a prognostic biomarker in patients with pancreatic cancer. Cancer Manag. Res. 2021, 13, 3675–3683.
- Yang, J.; Zhang, Y. Plasma-derived exosomal ALIX as a novel biomarker for diagnosis and classification of pancreatic cancer. Front. Oncol. 2021, 11, 628346.
- Hong, C.S.; Muller, L. Plasma exosomes as markers of therapeutic response in patients with acute myeloid leukemia. Front. Immunol. 2014, 5, 160.
- Wang, Y.; Niu, X. Exosomal PD-L1 predicts response with immunotherapy in NSCLC patients. Clin. Exp. Immunol. 2022.
- Momen-Heravi, F. Isolation of extracellular vesicles by ultracentrifugation. Methods Mol. Biol. 2017, 1660, 25–32.
- Merchant, M.L.; Powell, D.W. Microfiltration isolation of human urinary exosomes for characterization by MS. Proteomics Clin. Appl. 2010, 4, 84–96.
- Baranyai, T.; Herczeg, K. Isolation of exosomes from blood plasma: Qualitative and quantitative comparison of ultracentrifugation and size exclusion chromatography methods. PLoS ONE 2015, 10, e0145686.
- Zhu, L.; Sun, H.T. Isolation and characterization of exosomes for cancer research. J. Hematol. Oncol. 2020, 13, 152.
- Shen, M.; Di, K. Progress in exosome associated tumor markers and their detection methods. Mol. Biomed. 2020, 1, 3.
- Ramirez, M.I.; Amorim, M.G. Technical challenges of working with extracellular vesicles. Nanoscale 2018, 10, 881–906.
- Liu, C.; Zeng, X. Sensitive detection of exosomal proteins via a compact surface plasmon resonance biosensor for cancer diagnosis. ACS Sens. 2018, 3, 1471–1479.
- Carney, R.P.; Hazari, S. Multispectral optical tweezers for biochemical fingerprinting of CD9-positive exosome subpopulations. Anal. Chem. 2017, 89, 5357–5363.
- Liu, C.; Guo, J. Field-free isolation of exosomes from extracellular vesicles by microfluidic viscoelastic flows. ACS Nano 2017, 11, 6968–6976.
- Hosseini, R.; Asef-Kabiri, L. The roles of tumor-derived exosomes in altered differentiation, maturation and function of dendritic cells. Mol. Cancer 2021, 20, 83.
- Datta, A.; Kim, H. Manumycin A suppresses exosome biogenesis and secretion via targeted inhibition of Ras/Raf/ERK1/2 signaling and hnRNP H1 in castration-resistant prostate cancer cells. Cancer Lett. 2017, 408, 73–81.
- Datta, A.; Kim, H. High-throughput screening identified selective inhibitors of exosome biogenesis and secretion: A drug repurposing strategy for advanced cancer. Sci. Rep. 2018, 8, 8161.
- Im, E.J.; Lee, C.H. Sulfisoxazole inhibits the secretion of small extracellular vesicles by targeting the endothelin receptor A. Nat. Commun. 2019, 10, 1387.
- Catalano, M.; O’Driscoll, L. Inhibiting extracellular vesicles formation and release: A review of EV inhibitors. J. Extracell. Vesicles 2020, 9, 1703244.
- Menck, K.; Sönmezer, C. Neutral sphingomyelinases control extracellular vesicles budding from the plasma membrane. J. Extracell. Vesicles 2017, 6, 1378056.
- McKelvey, K.J.; Powell, K.L. Exosomes: Mechanisms of uptake. J. Circ. Biomark. 2015, 4, 7.
- Kirchhausen, T.; Macia, E. Use of dynasore, the small molecule inhibitor of dynamin, in the regulation of endocytosis. Methods Enzymol. 2008, 438, 77–93.
- Hayatudin, R.; Fong, Z. Overcoming chemoresistance via extracellular vesicle inhibition. Front. Mol. Biosci. 2021, 8, 629874.
- Christianson, H.C.; Svensson, K.J. Cancer cell exosomes depend on cell-surface heparan sulfate proteoglycans for their internalization and functional activity. Proc. Natl. Acad. Sci. USA 2013, 110, 17380–17385.
- Atai, N.A.; Balaj, L. Heparin blocks transfer of extracellular vesicles between donor and recipient cells. J. Neurooncol. 2013, 115, 343–351.
