Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Neurosciences
There are only a few techniques available that allow for the direct sampling from the cerebral interstitial space and thus provide insight into real concentrations in the brain parenchyma. Such techniques are microdialysis, cerebral open flow microperfusion (cOFM), and biosensors. Innovations in the field are discussed, along with the ‘nuts and bolts’ of the techniques.
- microdialysis
- cerebral open flow microperfusion
- electrochemical biosensors
- macromolecules
1. Introduction
Gaining insights into the pharmacokinetic and pharmacodynamic properties of lead compounds is crucial during drug development processes. When it comes to the treatment of brain diseases, collecting information at the site of action is challenging. Cerebrospinal fluid (CSF) concentrations do not necessarily reflect the real concentration in the brain parenchyma. Drug concentrations in the CSF give information regarding drug transport across the choroid plexus, which is the main component of the blood–CSF barrier, but such concentrations do not provide information concerning blood–brain barrier (BBB) transport. This misconception hinders progress in the development of drugs targeting the central nervous system (CNS), as explained by Pardridge [9]. Moreover, in recent years, the discovery of the glymphatic system, acting as a clearance pathway in the brain, stirred the debate about brain fluid dynamics even more [10,11,12]. Not only the influx but also the efflux mechanisms importantly impact the brain concentrations of (macro)molecules [13,14].
In fact, there are only a few techniques available that allow for the direct sampling from the cerebral interstitial space and thus provide insight into real concentrations in the brain parenchyma. Such techniques are microdialysis, cerebral open flow microperfusion (cOFM), and biosensors. While these techniques seem promising, they are not (yet) adopted into routine practice. The juxtaposition of these three techniques will lead to a more comprehensive overview of recent developments and possibilities in this domain.
2. Microdialysis
Microdialysis enables the continuous sampling of endogenous as well as exogenous compounds from the cerebral interstitial space using a probe with a semipermeable membrane. The probe is stereotactically implanted in the brain in the region of interest. A perfusion fluid, mimicking the ISF, often called artificial CSF, is perfused through the probe assembly using a controlled pulse-free syringe pump. At the outlet, the dialysate containing the substances of interest is collected without the need of a pull pump. The dialysate is collected fractionally and subsequently analyzed using a sensitive analytical method of choice. The underlying process for the exchange of substances is based on Fick’s first law of passive diffusion. In addition to the concentration gradient and osmotic pressure, the molecular weight, hydrophobicity, and tertiary structure of the compound, as well as the cut-off and material of the membrane, play key roles in this process [27].
The foundation for the use of microdialysis in its present form, as described above, originates from the early 1960s. The first building blocks were laid by Gaddum [28] by the introduction of a push-pull cannula to collect substances directly from the brain. His work is based on a perfusion technique in subcutaneous tissue described by Fox and Hilton [29], although the cannula was positioned concentrically in the brain tissue to allow for more precise targeting. The development of this in vivo technique evolved from the different attempts to determine neurotransmitters by performing brain dissections followed by post-mortem analyses. Numerous technical problems surrounded these early experiments, such as inaccurate dissections, the validity and correlation of the measurements in post-mortem tissue to the in vivo values, and the fact that only a single measurement of a static moment could be determined [30]. Over the years, it became clear that the in vivo technique had limitations as well that led to numerous adaptations regarding the design of the push-pull cannula. The major bottleneck was the open flow system resulting in tissue damage [31]. To resolve this problem, a cannula was constructed containing a tip covered with a porous semipermeable membrane. This dialysis sac [32], or ‘dialytrode’ as it was called by Delgado et al. [33], was later replaced by a hollow fiber, namely the dialysis membrane [34]. This is the basic principle underlying microdialysis as still referred to nowadays. The major advantage of this innovation is that there is less damage and interference with the brain tissue as exposure of the brain tissue to the perfusate is avoided, making the technique more ‘physiological’ than the push-pull principle [31,34,35].
The main component of a microdialysis probe is its semi-permeable membrane. As brain microdialysis was historically applied to gain insight into neurotransmitter levels and other small molecules, cut-off values of the membrane typically ranged from 6 to 40 kDa. Interestingly, the molecular weight cut-off (MWCO) of the microdialysis probe does not reflect the actual pore size of the membrane. It gives information regarding the retention capabilities and thus the sampling efficiency for molecules of a certain size range. For example, a membrane with a 20 kDa MWCO will not allow 80–90% of molecules of that particular size to pass through [36]. Furthermore, there is an exponential decrease in the ability of molecules to pass the semipermeable membrane in relation to an increase in their molecular weight, making classical microdialysis even challenging for sampling molecules with a low molecular weight because of its low recovery rates and low dialysate concentration. Over the years, effort was put in developing probes with a higher cut-off to increase the utility of the technique [15,37,38,39]. At present, probes with cut-off values of 100 kDa–3 MDa are commercially available, allowing the exchange of macromolecules [37,38]. In these kinds of probes, the underlying process for the exchange of substances is primarily based on convection, meaning substances are carried across the membrane pores via bulk-flow together with the solution. Ultrafiltration and, thus, transmembrane pressure (hydrostatic pressure gradient across the membrane) are crucial in this process [39]. To control the fluidic path, thus preventing leakage of the perfusate into the brain parenchyma, a push-pull system is required (Figure 1a). Nevertheless, pressure fluctuations remain a hurdle. Takeda et al. circumvented this problem by introducing a vent hole at the head of the probe assembly (AtmosLM™, Eicom, Green Leaf Scientific, Dublin, Ireland). This vent hole allows for fast equalization of the pressure difference inside the probe with the atmospheric pressure [40].
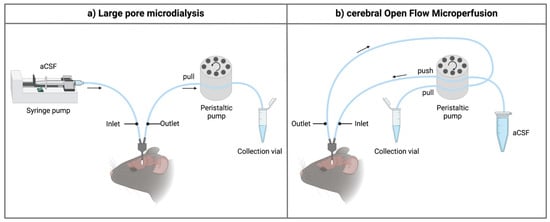
Figure 1. Schematic overview of the inlet and outlet tubings for the sampling techniques. (a) When using high-molecular-weight-cut-off microdialysis probes, a push-pull system is required to prevent loss of perfusion fluid through the large pores of the membrane. The setup generally contains a separate controlled pulse-free syringe pump and a peristaltic pull pump (e.g., using an AtmosLM™ or CMA ultra-high cut-off probe). For classical microdialysis, a pull pump is not used. (b) For the cerebral open flow microperfusion probe, a peristaltic push-pull microperfusion pump (MPP102 PC, Basi) can be used. Hereby, inflow and outflow can be controlled via the same pump head. Figure created with BioRender.com accessed on 6 April 2022. aCSF: artificial cerebrospinal fluid.
Samples obtained with classic microdialysis do not require sample clean-up before analysis with liquid chromatography or capillary electrophoresis, as no large molecules are present, because of the low MWCO of the membrane. Because of the high MWCO, analysis of large pore microdialysis samples is generally more challenging [38].
Technically, the microdialysis probe construct consists of a guide cannula containing a healing dummy implanted in the brain above the region of interest. Before initiating the sampling experiments, the healing dummy is removed and replaced by the microdialysis probe. The membrane protrudes beneath the guide cannula as illustrated in Figure 2a.
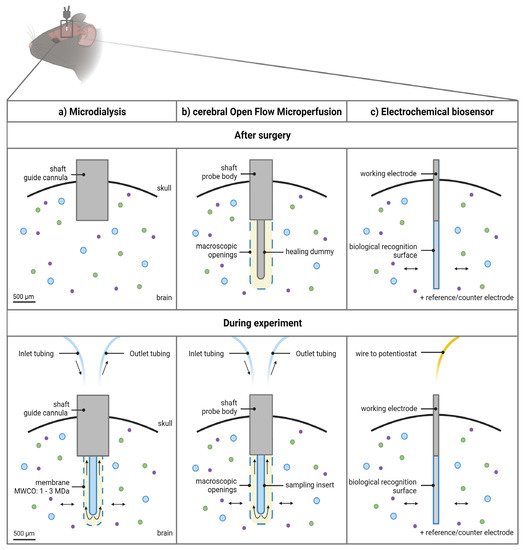
Figure 2. Schematic overview of the 3 approaches to monitor macromolecules directly from the cerebral interstitial fluid. (a) The microdialysis probe construct consists of a guide cannula containing a healing dummy implanted in the brain above the region of interest. Before initiating the experiments, the healing dummy is replaced by the probe connected to the tubings. The membrane protrudes beneath the guide cannula. Dimensions of the outer diameter of the shaft of the guide cannula and probe membrane are based on an AtmosLM™ probe. (b) The cerebral open flow microperfusion probe body construct/guide itself contains the open exchange area with macroscopic openings and is implanted directly into the brain in the region of interest. Before initiating the experiments, the healing dummy is replaced with the sampling insert connected to the tubings. (c) The electrochemical biosensor setup consists of the working electrode (cylinder type is shown), which is implanted in the brain region of interest, while the reference electrode can be implanted in the cortex and the counter electrode can be attached to an anchor screw placed in the skull (not shown). Scale bar indicates 500 µm. Figure created with BioRender.com accessed on 6 April 2022. MWCO: molecular weight cut-off.
3. Cerebral Open Flow Microperfusion
Quite soon after its first introduction in the 1960s, the push-pull perfusion technique with its open flow system was put aside. As the use of a membrane has shown its limitations as well (see Section 3), Birngruber et al. went back to the roots and introduced in 2013 an advanced technique referred to as cOFM [41]. The two main features of the patented cOFM probe body design itself are (i) the replacement of the membrane by macroscopic openings and (ii) the biocompatible material (fluorinated ethylene propylene) it consists of [42]. The open structure of the device allows for sampling lipophilic (these tend to adsorb to the microdialysis membrane) and high-molecular-weight substances. The biocompatible material should make it possible to perform chronic sampling experiments as will be discussed in detail in Section 3.1 [43]. Historically, one of the main hurdles to prevent tissue damage by using the push-pull technique was to maintain the probe inlet flow generated by the push pump equal to the probe outlet flow generated by the pull pump [30,31]. This problem was solved by using a pair of high-precision syringe pumps [42]. These were later replaced by a peristaltic microperfusion pump (MPP102 PC, Basi) where inflow and outflow can be controlled via the same pump head (Figure 1b) [25,44].
Technically, the membrane-free cOFM probe body construct itself contains the open exchange area necessary to perform the sampling experiments and is implanted directly into the brain in (not above, as for the microdialysis guide) the region of interest, as illustrated in Figure 2b. It is recommended by the manufacturer to implant the probe body 14 days before initiating the sampling experiments [45]. This recovery period should ensure re-establishment of the BBB (for detailed discussion, see Section 3.1.1) and can be appraised as a third major feature of the probe design [41,42]. A healing dummy prevents tissue growing into the probe during this period. Sampling experiments can be initiated after 14 days by replacement of the healing dummy with a sampling insert [45].
4. Electrochemical Biosensors
While the two above-mentioned techniques rely on fluid sampling, a third technique, electrochemical biosensors, depends on the recognition of certain molecules directly in the cerebral ISF [46]. The principle of an electrochemical biosensor was first described by Clark and Lyons, in 1962 [47]. A typical biosensor consists of an immobilized, biological recognition element combined with a transducer that converts the biological reaction to a quantifiable signal. The biological recognition element is immobilized on the electrode surface. An electrochemical biosensor uses an electrochemical transducer to convert the signal, which is an electrical current in the case of an amperometric biosensor [48,49,50]. The measured electrical signal is a result of the redox current that is present at the surface of the electrode [51,52]. In this work, the focus is on electrochemical biosensors as this type of biosensor is not only used in fundamental scientific research [53] but also used in clinical practice. For example, the glucose biosensor has been used by diabetic patients for continuous glucose monitoring [50].
The first electrochemical biosensors were developed for the biosensing of molecules by the use of enzymes [50]. Enzymatic biosensors can be divided into three generations. In first-generation biosensors, an enzyme is used as the recognition element and the products used or produced in the enzymatic reaction are in relation to the concentration of the analyte of interest. Second-generation biosensors use electron carriers, while third-generation biosensors do not depend on a mediator but instead use direct electron transfer [51,52]. Enzymatic biosensors still make up the majority of implantable biosensors [54,55]. Apart from enzymes as the biological recognition element, aptamers, antibodies, or antigens can also be used [49,56] to (specifically) recognize the analyte of interest. Nevertheless, interference can be expected by other (small) electroactive compounds. A selective membrane layer/polymer is often used to prevent interference [57,58].
The best examples for the in vivo use of this elegant technique are the glucose biosensor [50] and the assessment of several endogenous small brain molecules such as glutamate [53,59,60] or other molecules [61,62]. While the use of biosensors to establish drug concentrations in the brain is limited, there are examples of biosensors that have been developed for the determination of disease biomarkers [63,64,65] useful for personalized medicine [23,26,66]. Nowadays, biosensors are also being developed for the measurement of large molecules, such as for the detection of tau protein as the biomarker for neurodegenerative diseases [63] or for the detection of hepatitis C virus for diagnostic purposes [67,68,69]. However, the latter are mostly being developed for ex vivo use in biological samples and not yet routinely in vivo. For example, several groups have described the development of a tau protein biosensor, of which an overview can be found in the review by Ameri et al. [20]. The aggregation of tau protein is associated with neurodegeneration and Alzheimer’s disease [17,63]. While the aggregated tau is usually seen in later stages of the disease, it can be of interest to identify soluble tau oligomers during the early stages of the disease. The applicability of biosensors for this purpose is shown in an in vitro setting by Esteves-Villanueva et al. [63]. Another example is the development of an electrochemical biosensor for the determination of the HER2 receptor in cell or tumor lysates, which is the target of the aforementioned trastuzumab monoclonal antibody against cancer and requires characterization of HER2 [26,66].
Typically, in an in vivo setting, a three-electrode system is used: (i) the working electrode (which is the biosensor), (ii) the reference electrode, and (iii) a counter electrode [67,69,70]. Specifically, to monitor extracellular concentrations in the brain tissue, the working electrode(s) is (are) implanted in the brain region of interest (Figure 2c), while the reference electrode can be implanted in the cortex and the counter electrode can be attached to an anchor screw placed in the skull [71].
This entry is adapted from the peer-reviewed paper 10.3390/pharmaceutics14051051
This entry is offline, you can click here to edit this entry!
