Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Gastroenterology & Hepatology
Inflammatory bowel disease (IBD) is a chronic inflammatory disorder of the gastrointestinal tract. It is characterized by relapses and remissions, thus requiring lifelong treatment.
- BD
- risk factors of IBD
1. Introduction
Inflammatory bowel disease (IBD) is a chronic inflammatory disorder of the gastrointestinal tract. It is characterized by relapses and remissions, thus requiring lifelong treatment. The etiology of the disease is complex. Despite significant advances in the molecular biology and knowledge regarding IBD, the pathogenesis of the disease remains unclear. Since the microbial community has been found to play a huge role in the human health and has been proved to be altered in IBD patients, it seems that it could be one of the crucial elements of IBD development. In addition to the already well demonstrated immunological, genetic and environmental risk factors of IBD.
2. Inflammatory Bowel Disease
Inflammatory bowel diseases (IBD), which include ulcerative colitis (UC) and Crohn’s disease (CD), are among disorders with still undetermined etiology. They comprise numerous immunological, genetic, microbiological, environmental and dietary factors [1]. In recent years, progress has been observed in the search for the genetic factors associated with IBD. In fact, it has been proved that one of the elements affecting the functions of the immune system are genetic mutations responsible for the expression of numerous proteins that have a regulatory effect. However, studies of monozygotic twins demonstrate low compliance rates in the incidence of these diseases, in CD (20–55%) and UC (6.3–17%), which in turn confirms the hypothesis that other coexisting factors must be involved. More than 200 genes have been discovered that predict the development of IBD. Most of them are genetic polymorphisms associated with the function of the mucosal barrier, responsible for the direct interaction with gastrointestinal antigens, including the microbiota and dietary components [2,3,4,5]. Furthermore, an epidemiological study has indicated that significantly more cases of IBD are observed in highly developed countries. It is also worth noting that first-generation immigrants who had moved to these countries were at a higher risk of IBD [6]. These observations confirm the undeniable influence of environmental factors on the development of the afore-mentioned diseases, and they impact the gastrointestinal microbiota together with dietary factors. Nowadays, the microbiota is seen as an integral part of keeping the human body healthy. A microbiome is a collection of microbes genes in the host organ [7]. In many investigations, it has been proved that disorders in the composition of the intestinal microbiota, defined as a complex of microorganisms inhabiting the intestines, directly affect the cells of the immune system, thus stimulating its activity and inducing the activity of inflammation in the mucosal membrane. In fact, some reports indicate that patients with IBD present a characteristic dysbiosis [8,9]. The role of the intestinal microbiota in the pathogenesis of IBD may also be confirmed by the effect of antibiotic therapy, probiotic intake and the beneficial effect of fecal microbiota transplantation on the induction of disease remission [10]. The most relevant findings related to IBD are shown in Figure 1 (based on Mulder et al.) [11].
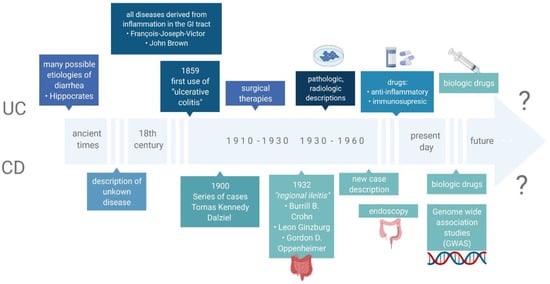
Figure 1. The most relevant findings related to IBD.
Although diet seems to play a significant role in the pathogenesis of IBD, epidemiological studies regarding its impact on these disorders are challenging to perform due to a large number of confounding factors. However, based on the research conducted to date, it can be concluded that a specific nutritional profile has a beneficial effect on the course of IBD. An increased intake of unsaturated fatty acids, fruits, vegetables and breastfeeding undoubtedly shows a protective effect on the intestines. Recently, much attention has been paid to the Mediterranean diet, and it seems that its promotion contributes to reducing the risk of developing IBD [12,13].
3. Risk Factors of IBD
3.1. Genetics
The basic genetic knowledge of the IBD hereditary character stems from comparative studies that investigated the prevalence of the disorder in first-degree family members, relatives of the affected individuals, as well as from twin studies. In 2002, research results conducted on 1000 IBD patients over the period of 20 years were described [14]. As many as 14% of respondents reported at least one family member in the first line with a CD in their family history. This is consistent with the studies conducted by other researchers who estimate the group of CD patients in the family history at 10–20% [15]. Having a first-degree relative family member diagnosed with CD increases the risk of developing the disease 15–35 times, whereas, in the same circumstances, the risk for UC is increased only 6–9 times [16]. Observations of monozygotic twins with CD proved that the incidence rate is 20–58.3% (average 30.3%). In fraternal twins, it does not differ from the incidence observed in non-twin siblings and is equal to approximately 3.6%. In terms of UC, the correlation is much weaker and amounts to 6.3–18.2% (average 15.4%) [17,18,19].
The first study indicating the locus associated with CD development was published in 1996 by Hugot and co-authors. The location at the band 12 in the short arm (q) of chromosome 16 (16q12) was then defined as the IBD1 region [20]. In 2001, the same research team achieved another success by narrowing down the IBD1 area to a specific NOD2 gene and three major mutations: rs2066844 (c.2104C > T, p.Arg702Trp in exon 4), rs2066845 (c.2722G > C, p.Gly908Arg in exon 8) and rs2066846 (c.3019_3020insC, p.Leu1007fs and in exon 11) associated with CD diagnosis within this gene [21,22] (Figure 2). In fact, these variants may increase the risk of developing the disease up to 40 times [23]. The NOD2 gene (formerly known as CARD15) encodes an intracellular receptor that regulates the non-specific immune response, expressed in peripheral blood monocytes, macrophages and intestinal mucosa cells, particularly Paneth cells. The NOD2 protein, consisting of two N-terminal domains (CARD), a nucleotide-binding domain (NBD) and a C-terminal end containing ten leucine-rich repeats (LRR), is involved in bacterial pathogen recognition and inflammasome initiation via binding to muramyl dipeptide. To date, NOD2 represents the best-known gene with regard to the involvement of genetic factors in CD [24]. The recessive inheritance of rare and low-frequency deleterious NOD2 variants accounts for 7–10% of the CD cases [25]. Moreover, the relationship between NOD2 mutations and the location of the disease in the ileum area has been proved. However, differences in the occurrence of the mutations in the region of this gene have been observed, e.g., the Japanese, Chinese, and Koreans are the populations free from NOD2 variants, whereas the changes occur very rarely in the African Americans suffering from IBD [26,27].
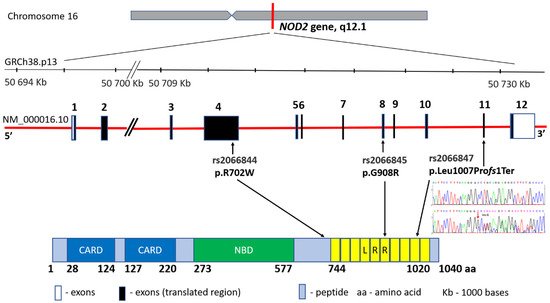
Figure 2. NOD2 gene structure, main variants’ distribution and the protein product.
According to data from the Online Mendelian Inheritance in Man (OMIM) database, to date 28 regions (IBD1-28) have been defined in the human genome containing specific genes and changes that are associated with an increase in IBD incidence (Table 1).
Table 1. IBD chromosome regions and main candidate genes.
| Region Name | OMIM Number |
Location | Disease | Marker (LOD Score), rs Number (p-Value) |
Candidate Genes |
|---|---|---|---|---|---|
| IBD1 | 266,600 | 16q12 | CD | D1S3669 (2.65) | NOD2 |
| IBD2 | 601,458 | 12p13.2-q24.1 | CD, UC | D12S83 (5.47) | VDR, IFNG |
| IBD3 | 604,519 | 6p21.3 | CD, UC | D6S289 and D6S276 (2.07) D6S461 (4.2) |
HLA-B, HLA-DRB1, TNF, LTA |
| IBD4 | 606,675 | 14q11-q12 | CD | D14S261 (3.0) | RNASE2, RNASE3 |
| IBD5 | 606,348 | 5q31-q33 | CD | 5q31-q33 (3.9) | IL4, IL5, IL13, IRF1, CSF2, SLC22A4, SLC22A5 |
| IBD6 | 606,674 | 19p13 | CD, UC | D19S591 (4.6) | ICAM1, C3, TBXA2R, LTB4H |
| IBD7 | 605,225 | 1p36 | CD, UC | D1S1597 (3.01) | TNFRSF1B, TNFRSF4 CASP9 |
| IBD8 | 606,668 | 16p | CD, UC | D16S408 (>2.5) | IL27, SULT1A1, SULT1A2 |
| IBD9 | 608,448 | 3p26 | CD, UC | D3S1297 (3.69) | |
| IBD10 | 611,081 | 2q37.1 | CD | ATG16L1 | |
| IBD11 | 191,390 | 7q22 | CD, UC | D7S669 (3.08) | MUC3A |
| IBD12 | 612,241 | 3p21.3 | CD, UC | D3S2432 (1.68) | MST1, BSN, GNAI2 |
| IBD13 | 612,244 | 7q21.1 | CD, UC | D7S669 (3.08) | ABCB1 |
| IBD14 | 612,245 | 7q32.1 | CD, UC | IRF5 | |
| IBD15 | 612,255 | 10q21 | CD, UC | rs224136 (<10 × 10−10) | SIRT1 |
| IBD16 | 612,259 | 9q32 | CD, UC | D9S2157 (1.41) | TNFSF15 |
| IBD17 | 612,261 | 1p31.3 | CD, UC | rs11209026 (<10−9) | IL23R |
| IBD18 | 612,262 | 5p13.1 | CD, UC | rs1373692 (2.1 × 10−12) | PTGER4 |
| IBD19 | 612,278 | 5q33.1 | CD | IRGM | |
| IBD20 | 612,288 | 10q23-q24 | CD, UC | D10S547 and D10S20 (2.30) | DLG5 |
| IBD21 | 612,354 | 18p11 | CD, UC | rs2542151 (3.16 × 10−8) | PTPN2 |
| IBD22 | 612,380 | 17q21.2 | CD | rs744166 (6.82 × 10−12) rs2872507 (5.00 × 10−9) |
STAT3, ORMDL3 |
| IBD23 | 612,381 | 1q32.1 | CD, UC | rs11584383 (1.43 × 10−11) | IL10 |
| IBD24 | 612,566 | 20q13 | CD, UC | rs2315008 (6.30 × 10−8) rs4809330 (6.95 × 10−8) | TNFRSF6B |
| IBD25 | 612,567 | 21q22.1 | CD, UC | rs2836878 (6.01 × 10−8) | IL10RB |
| IBD26 | 612,639 | 12q15 | UC | rs1558744 (2.5 × 10−12) | |
| IBD27 | 612,796 | 13q13.3 | CD | rs20411 (LOD 3.98) | |
| IBD28 | 613,148 | 11q23.3 | CD, UC | IL10RA |
ABCB1: ATP binding cassette subfamily A member 1; ATG16L1: autophagy related 16 like 1; BSN: bassoon presynaptic cytomatrix protein; CASP9: caspase 9; CD: Crohn’s disease; C3: complement C3; CSF2: colony stimulating factor 2; DLG5: discs large MAGUK scaffold protein 5; GNAI2: G protein subunit alpha i2; HLA-B: major histocompatibility complex, class I, B; HLA-DRB1: major histocompatibility complex, class II, DR beta 1; ICAM: intercellular adhesion molecule, IFNG: interferon gamma; IL4, IL5, IL10, IL13 and IL27: interleukins 4, 5, 10, 13 and 27, respectively; IRF1 and IRF5: interferon regulatory factors 1 and 5, respectively; IL10RA and IL10RB: interleukin 10 receptor subunits alpha and beta, respectively; IRGM: immunity-related GTPase; LOD: logarithm (base 10) of odds; LTA: lymphotoxin alpha; LTBH4H: leukotriene B4 hydroxylase; MST1: macrophage stimulating 1; MUC3: mucin 3; NOD2: nucleotide binding oligomerization domain containing 2; RNASE2: ribonuclease A family member 2; RNASE3: ribonuclease A family member 3; ORMDL3: ORM1-like protein 3; PTPN2: protein tyrosine phosphatase, non-receptor type 2; SLC22A4 and SLC22A5: solute carrier family 22 members 4 and 5, respectively; STAT3: signal transducer and activator of transcription 3; SULT1A1 and SULT1A2: sulfotransferase 1A members 1 and 2, respectively; TBXA2R: thromboxane A2 receptor; TNF: tumor necrosis factor; TNFRSF1B and TNFRSF4: TNF receptor superfamily members 1B and 4, respectively; UC: ulcerative colitis; VDR: vitamin D receptor.
The list of DNA loci that may be related to the pathogenesis of IBD is much longer and includes nearly 300 candidate genes, mainly as a result of genome-wide association studies (GWAS) [28,29,30,31]. In addition to the DNA sequence variants, epigenetic research is also a new, promising research area in IBD. DNA modifications appear to be causally involved in the pathogenesis of IBD, as evidenced by studies of CD8 + T-cell in vitro cultures obtained from IBD patients [32], or genome-wide analysis of the DNA methylation profile [33]. Such epigenetic research opens up a broad field of research that generates exciting insights into IBD pathophysiology. The patterns of DNA methylation and histone modification as well as microRNA (miRNA) molecules post-transcriptional regulation may in the future serve not only as biomarkers of disease predisposition, disease activity, or disease course, but also as new targets in therapeutic interventions in IBD patients [34,35].
The genetic factors involved in IBD represent a comprehensive issue. In this paper, which focuses on dietary patterns and microbiota in IBD patients, it is worth emphasizing that the subject of recent research is genetic defects affecting the homeostasis of the intestinal barrier. In particular, the impaired expression of genes related to the cellular involvement, junction complexes, mucus production and secretion, pathogen detection, Paneth cell activity, reactive oxygen species production, xenobiotic responseand IgA secretion severely impairs the intestinal epithelial barrier integrity and protective functions in IBD patients [36]. For instance, studies on a genetically engineered mouse model have shown that the dysregulated expression of genes encoding commitment-related transcription factors, such as SRY-box transcription factor 9 (Sox9), hairy and enhancer of split 1 (Hes1), serine-threonine kinase 11 (Stk11), mouse atonal homolog 1 (Math1), caudal type homeobox 2 (Cdx2) and GATA-binding factor 6 (Gata6), definitively compromises intestinal epithelial cell differentiation and, as a consequence, intestinal barrier integrity with its secretive (particularly Paneth cells) and adhesion functions [37,38,39,40]. Moreover, in Crohn’s disease patients, the impairment of epithelial junctional complexes was found to significantly contribute to the development of chronic inflammatory conditions, which may be caused by an increased expression of claudin-2 (CLDN2) and an impaired expression and redistribution of claudin-5 (CLDN5), claudin-8 (CLDN8) and occludin (OCLN). Hence, an increase in intestinal permeability and bacterial translocation is observed [41]. Similar relationships have been described in the studies of ulcerative colitis patients in whom the deregulated expression of OCLN, CLDN1, junctional adhesion molecule 1 (F11R, also known as JAM), beta-catenin (CTNNB1) and epithelial cadherin (CDH1) led to the transepithelial migration of neutrophils [42]. Moreover, animal studies confirm these observations, e.g., that mice with a knockout of the F11r gene, despite having a normal structure of the intestinal epithelium, developed low-grade colonic inflammation [43].
O-glycans of the gastrointestinal tract constitute the first defense line against external stimuli. Defects in O-glycosylation may lead to damage in mucin expression classified into three categories: secretory gel-forming (MUC2, MUC5AC, MUC5B and MUC6), secretory non-gel-forming (MUC7) and membrane-bound (e.g., MUC1, MUC3, MUC4, MUC12, MUC13 and MUC17). Consequently, mucosal barrier destruction occurs, leading to the microbial activation of inflammasomes [44]. Particularly, Helicobacter pylori and Clostridium difficile infections are associated with disrupted mucin synthesis and mucus barrier, whereas Helicobacter pylori-infected patients present with a significant decrease in MUC5AC gene expression levels [45].
In conclusion, it should be noted that a new area in the study of genetic factors of IBD has recently been established. Haberman et al., in their observations, demonstrated a significant reduction in the expression of mitochondrial genes encoding the oxidative phosphorylation chain and nuclear genes in patients with UC [46]. Researchers have suggested and simultaneously confirmed in preliminary studies that mitochondriopathy is also a process associated with the pathogenesis of IBD [47].
3.2. Immunology
Research with regard to intestinal immunology has long played a fundamental role in understanding the pathogenesis of IBD and has primarily focused on investigating the mucosal response of the intestines. It has been suggested that disorders in innate immunity, as well as adaptive immunity, contribute to the development of IBD.
The adaptive response is specific and continues for several days; it is also predominantly T-lymphocyte dependent. Most researchers investigating this type of response favor the theory that CD is a disease associated with increased Th1 lymphocyte activity, while UC entails the activity of Th2 lymphocytes [48,49]. Th1 cells produce large amounts of IFNγ, IL-2 and TNFα, and Th2 cells release IL-4, IL-5 and IL-13. Some studies have indicated that, in cells grown in vitro for both CD and UC, exact high amounts of IFNγ are observed [50]. Furthermore, research conducted in recent years has suggested that Il-13 has anti-inflammatory effects [51]. Wilson et al. observed that supernatants from biopsies grown in vitro for both UC and CD have lower IL-13 concentrations compared to IFNγ. In addition, researchers have found that the concentrations of these cytokines are comparable in both diseases [52]. Therefore, some researchers believe that both forms of Th-lymphocyte response coexist in patients with IBD. In terms of adaptive response, it is essential to take into consideration the Th17 cell. High concentrations of IL-17A, produced by these lymphocytes, were found in both the mucous membrane of patients with CD and UC in comparison to the control group. In fact, Th17 cells also constitute an important source of IL-21, a cytokine associated with the activity of IL-2, one of the primary inflammatory cytokines in the intestine. It has also been shown that there is an imbalance in IBD between Th17 lymphocytes and regulatory T (Treg) lymphocytes. Th17 cells activate the inflammatory process, and Treg cells inhibit autoimmunity in IBD. To maintain the proper balance of these cells, factors that stimulate cytokine signaling, such as intestinal microflora or bile acid metabolites, also play a role [53]. Much attention has also been paid to innate forms of the immune response, such as the integrity of the epithelial barrier, the reaction to the intestinal microbiota, and food antigens derived from its lumen. This form of response is our first line of defense against pathogens—thanks to it, unlike the adaptive response, the body reacts quickly to stimuli within minutes or hours. The first barrier to food and bacterial antigens coming from the intestinal lumen is its mucous layer. The second line is the intestinal epithelium, consisting of enterocytes and other specialized cells. In addition to mucus, these cells can produce a number of antimicrobial peptides. The defective expression has been proved in patients with CD [54]. The second line of defense consists of numerous cells, such as epithelial cells, macrophages, NK cells, monocytes and dendritic cells. Their activation occurs via Toll-like receptors (TLR) located on the surface of cells, and NOD-like, located in the cytoplasm. In patients with IBD, we observe the destruction of these receptors and effector cells [55]. A key cytokine that plays a role in the early response to microbial antigens is IL-23. Its polymorphisms are associated with both CD and UC, making it a typical cytokine in inflammation activity in IBD. IL-23 induces Th17 cells and cells of the innate immune system [56].
Many studies focus on mutations in the NOD-2 receptor range. These mutations are primarily associated with susceptibility to CD. Their consequence is an abnormal response of the immune cells of the intestine to bacterial lipopolysaccharides (LPS). There are various hypotheses as to the role of these mutations. It is believed that it may contribute to the lack of inhibition of TLR2 receptors, which results in the uncontrolled development of the inflammation and activation of Th-1 lymphocytes. Another theory is related to the reduced activation of NFkB, associated with a reduced reaction of the immune system cells to bacterial antigens [57].
An essential element of maintaining cellular homeostasis and an element of the host’s defense against foreign antigens is autophagy, which consists of the controlled decomposition by cells of chemical molecules, cell organelles and other cell fragments. The protein complex ATL16L1 (autophagy-related 16-like 1) is responsible for the proper phenomenon of autophagy. It has been shown that, in patients with IBD, there is a mutation in the ATL16L1 gene, which results in the excessive elimination of intestinal epithelial cells and, consequently, increased permeability [58].
Immunological factors associated with Crohn’s disease and ulcerative colitis are presented in Table 2.
Table 2. Immunological factors associated with Crohn’s disease and ulcerative colitis.
| Crohn’s Disease | Ulcerative Colitis |
|---|---|
| Th1 | Th2 |
| IFNγ, IL-2, TNFα | IL-4 |
| IL-2 | IL-5 |
| TNFα | IL-13 |
| Il-17A | |
| Th17 lymphocyte and regulatory T (Treg) lymphocyte imbalance | |
| Il-23 polymorphism | |
3.3. Environment
Several epidemiological and laboratory data demonstrate that the etiology of IBD is the result of an interplay of genetic, immunological and environmental factors. Environmental factors associated with IBD include smoking; post-appendectomy status; the use of oral contraceptives, antibiotics and non-steroidal anti-inflammatory drugs (NSAIDs), which may affect the composition of the gut microbiome; diet; breastfeeding; and past infections, vaccinations, and environmental pollution [59,60,61]. Unfortunately, the results of many studies on the role of environmental factors in the pathogenesis of IBD often remain inconsistent.
One of the best-studied factors is smoking. In his meta-analysis, Calkins showed that active smokers were less likely to develop ulcerative colitis compared to those who did not smoke or had stopped smoking. The study also showed that active and former smokers had a significantly increased risk of Crohn’s disease [62]. In addition, in patients with CD, smoking may be associated with an aggressive disease course, more surgical interventions and the need for intensified treatment [63,64]. It has also been shown that women with CD may be particularly adversely affected by smoking [65].
The exact mechanisms of the effects of tobacco smoke on the development of IBD are not well understood. One theory is that nicotine, via nicotinic acetylcholine receptors (nAChRs) present on T cells, may affect their function and cause the modulation of cellular immunity and the alteration of the cytokine profile [66]. Other hypotheses include altering the composition of the gut microbiome and generating oxidative stress due to free radicals [67].
Another important risk factor is having an appendectomy after the age of 20. It has been shown that the so-called early appendectomy is associated with a reduced risk of UC [68]. Data on the association of appendectomy with the development of CD remain conflicting. Frisch et al. and Koutroubakis et al. showed that appendectomy is a predisposing factor for CD [69,70], while Radford-Smith et al. report its protective role [71]. However, the mechanism of how appendicitis affects the risk of developing IBD is not yet fully understood. It has been postulated that bacteria in the appendix may play a role in the immune response to the intestinal microflora.
Khalili et al. found an association between oral contraceptive use and IBD development, particularly CD [72]. It has been suggested that the estrogens in these drugs may enhance the humoral response and stimulate macrophages; progesterone, on the other hand, has strong immunosuppressive effects [73].
The interaction between the gut microbiome and the intestinal barrier has been shown to play an important role in the pathogenesis of IBD. Thus, the use of drugs, such as antibiotics and non-steroidal anti-inflammatory drugs (NSAIDs), that modify the composition of the gut microbiota may contribute to an increased risk of disease [74]. Aniwan et al., in a population-based case–control study, showed a strong association between antibiotic use and the risk of CD as well as UC. This risk was increased for all forms of IBD according to age [75].
Antibiotic use during childhood may be particularly important. It has been postulated that it may lead to the impaired development of gut bacteria tolerance, which is a direct risk factor for IBD [76]. Ananthakrishnan et al. suggest that the frequent use of NSAIDs, but not aspirin, seemed to be associated with an increased absolute incidence of CD and UC [77]. NSAIDs may cause direct damage to the gastrointestinal mucosa and, through cyclooxygenase inhibition, affect prostaglandin production and thus exert important immunoregulatory functions.
The developmental hygiene hypothesis is also worth mentioning, which assumes that the increase in immunological disorders can be attributed to improved hygienic and sanitary conditions of the population and consequently less exposure to intestinal pathogens in childhood; it leads to an abnormal response to antigens later in life [78].
Water and air pollution associated with urbanization and industrial development may also contribute to the development of IBD. These include particulate matter, nitrogen dioxide, sulfur dioxide and pollutants from fossil fuel combustion, which can migrate across the gastrointestinal barrier and cause DNA damage and the disruption of immune mechanisms, which can result in the development of IBD [79].
Stress is also one of the important factors influencing the development of inflammatory bowel diseases [80,81]. It is well known that stress factors can affect immune functions via neuropeptides released from nerve cells, which in turn can lead to damage to the intestinal mucosa and thus impair its function as a protective barrier [82,83]. Bitton et al. suggest that individuals presenting lower stress levels show a reduced risk of Crohn’s disease [84]. Camara et al. emphasize above that the association between perceived stress and the exacerbation of Crohn’s was entirely related to mood components, specifically anxiety and depression [85].
It is worth mentioning that breastfeeding is a recognized protective factor for IBD. Human milk is believed to contain many substances that can affect growth and development and gastrointestinal function. In addition, the composition of the colonic bacterial flora has been shown to differ between breastfed and bottle-fed infants [86].
Understanding the influence of specific environmental factors on the development and course of IBD requires further detailed studies with different populations. Environmental factors influencing the risk of inflammatory bowel disease are presented in Table 3.
Table 3. Environmental factors influencing the risk of inflammatory bowel disease.
| Factors | Risk of Crohn’s Disease | Risk of Ulcerative Colitis |
|---|---|---|
| Smoking | ↑ | ↓ |
| Appendectomy over the age of 20 | Conflicting data | ↓ |
| Oral contraceptives | ↑ | |
| Antibiotics and non-steroidal anti-inflammatory drugs (NSAIDs) | ↑ | |
| Water and air pollution | ↑ | |
| Stress | ↑ | |
| Being breastfed | ↓ | |
↑—increase, ↓—decrease.
3.4. Gut Microbiota
Over one hundred trillion various microorganisms, including bacteria, archaea, fungi, viruses and protozoa, colonize the human gastrointestinal tract (GI), constituting the gut microbiota [87]. This complex ecosystem plays a fundamental role in the host organism. Gut-associated lymphoid tissue (GALT), as part of the host immune system, is involved in the intestinal immune and metabolic responses via fermentation products, such as short-chain fatty acid (SCFA) and acetate, also in the regulation of host mucosal homeostasis, hormones release and vitamin synthesis. Thus, its impact on human health is crucial [88,89]. The power of this ecosystem also supports the fact that the collective genome of gut microbial flora is estimated to contain 100 times more genes than the human genome [90].
The gut microbiota also influences the immune system, which has a direct effect on the maintenance of the intestinal immune barrier and the development of inflammatory bowel disease. Moreover, the impact of both commensal bacteria and pathological biota on the immune system has been emphasized. Commensal microorganisms stimulate the maturation and differentiation of Treg lymphocytes, and affect the production of IL-10, which plays a key role in maintaining Th1/Th2 homeostasis; thus, they affect the development of the immunotolerance and maintenance of cytokine balance.
CD4 + T lymphocytes of the intestinal wall plaque secrete IL-17 and IL-22, which have a regulatory function in the development of the inflammatory process in the intestine, whereas IL-17 induces the expression of the chemokines CXC and CC [91,92]. In turn, Peyer cells contribute to the production of B cells and plasma cells, which produce IgA, protective of the intestinal barrier [93,94].
In germ-free mice, the mRNA expression level of the bactericidal protein Ang4, secreted by Penatha cells, is significantly lower compared to conventional mice, which also confirms the involvement of biota in the intestinal mucosal immune barrier function [95].
Additionally, the microbiota affects the regulation of the CARD9 gene responsible for inflammation in IBD [96].
In patients with moderate colitis, IL-22 production was elevated following the exposure to altered gut microbiota [97].
In IBD, the amount of SCFAs produced by the intestinal biota is significantly reduced [98]. In fact, SCFAs affect the immune system by binding GPR43 and provides the homeostasis of colonic Treg cells [99,100].
Research conducted in recent years has shown that disturbances in the composition of the gut microbial community (dysbiosis) and in its metabolic balance as well as, finally, the interaction of microbiota with the host are associated with IBD pathogenesis [8,101,102,103]. Although it is still not fully known whether these changes cause the disease or, rather, are a consequence of the intestinal inflammation. However, there is evidence that some microbial metabolites may directly affect the inflammation process via the expression of genes, such as anti- and pro-inflammatory cytokines, and can influence T-helper cell (Th) 1, 2 and 17 and regulatory T-cell (Treg) release [104]. In particular, commensal bacteria from Clostridium clusters can induce the differentiation of colonic Treg cells by producing SCFA [105]. Studies performed on germ-free mice revealed a reduced development of Th17 cells and a decreased number of lymphocytes and immunoglobulin (Ig) A in the intestinal mucosa of these animals compared to controls [106,107,108,109].
Numerous animal studies confirm the contribution of the intestinal microbiome in IBD pathogenesis. Researchers observed that a germ-free environment prevents UC in genetically susceptible mice [110]. Nevertheless, the transfer of dysbiotic bacteria to the healthy mice induces and develops inflammation [111]. Furthermore, the colonization of mice with gut microbiota obtained from IBD patients alters the immune response and exacerbates UC in mice [112]. Additionally, the transfer of naive CD4+ lymphocytes from the healthy mice to those lacking T and B lymphocytes may induce UC, wherein the susceptibility depends on the gut microbiota composition [113].
Multiple observations of human subjects also confirm the contribution of the microbial community in the mechanism of IBD induction. It has been observed that IBD activity is most apparent in the parts of the intestine where bacterial populations are high and at a standstill (colon and rectum). In CD patients, disease remission occurs in the segment of the intestine excluded from the fecal stream. Similarly, the post-operative re-exposure to the fecal flow in the intestine correlates with CD relapse [114].
Although the IBD pathogenesis has not yet been fully understood, new insights continue to emerge through research. The disease is a complex interplay of the environment, immune system and microbiota in a genetically susceptible host [115]. Table 4 shows the most important changes in the gut microbiota in inflammatory bowel disease.
Table 4. The most important changes in gut microbiota in inflammatory bowel disease.
| Decreased Abundance | Increased Abundance |
|---|---|
| Firmicutes | Ruminococcus gnavu |
| Faecalibacterium prausnitzii | Proteobacteria |
| Lactobacillus | Actinobacteria |
| Roseburia faecis | Escherichia coli |
| Clostridium XIVa | Desulfovibrio |
| Clostridium IV | Akkermansia muciniphila |
| Faecalibacterium prausnitzii | Escherichia |
| Bacteroidetes | Fusobacterium |
| Verrucomicrobia | |
| Anaerostipes | |
| Methanobrevibacter | |
| Faecalibacterium | |
| Peptostreptococcaceae | |
| Collinsella | |
| Christensenellaceae |
This entry is adapted from the peer-reviewed paper 10.3390/nu14122520
This entry is offline, you can click here to edit this entry!
