Technological innovation has enabled the development of machine learning (ML) tools that aim to improve the practice of radiologists. In the last decade, ML applications to neuro-oncology have expanded significantly, with the pre-operative prediction of glioma grade using medical imaging as a specific area of interest.
- clinical implementation. Keywords: artificial intelligence
- glioma
- machine learning
- deep learning
1. Introduction
1.1. Artificial Intelligence, Machine Learning, and Radiomics
1.2. Machine Learning Applications in Neuro-Oncology
2. Algorithms for Glioma Grade Classification
3. Challenges in Image-Based ML Glioma Grading
3.1. Data Sources
3.2. External Validation
3.3. Glioma Grade Classification Systems
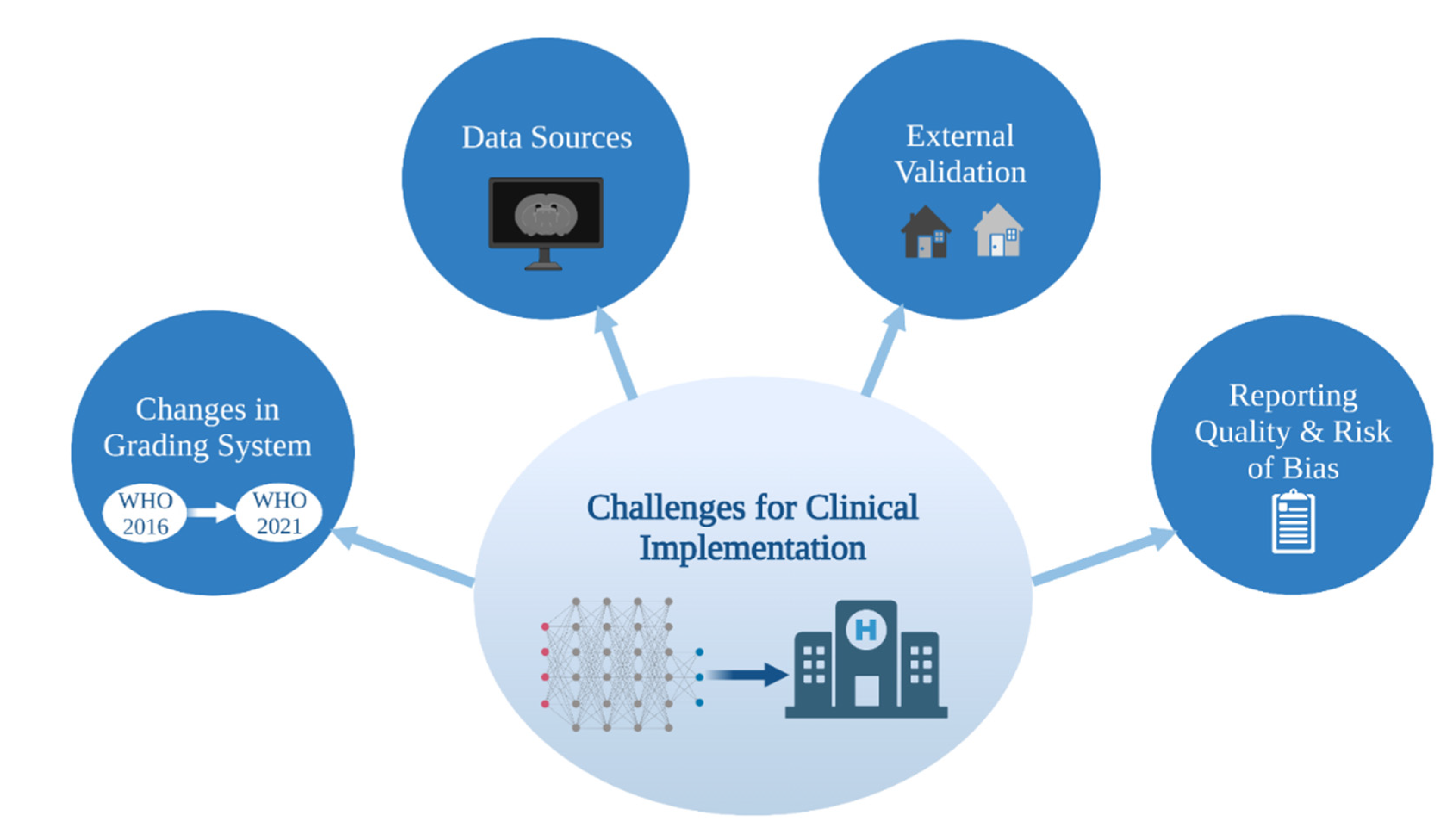
3.4. Reporting Quality and Risk of Bias
3.4.1. Overview of Current Guidelines and Tools for Assessment
Clear and thorough reporting enables more complete understanding by the reader and unambiguous assessment of study generalizability, quality, and reproducibility, encouraging future researchers to replicate and use models in clinical contexts. Several instruments have been designed to improve the reporting quality (defined here as the transparency and thoroughness with which authors share key details of their study to enable proper interpretation and evaluation) of studies developing models. The Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD) Statement was created in 2015 as a set of recommendations for studies developing, validating, or updating diagnostic or prognostic models [36]. The TRIPOD Statement is a checklist of 22 items considered essential for transparent reporting of a prediction model study. In 2017, with a concurrent rise in radiomics-based model studies, the radiomics quality score (RQS) emerged [37]. RQS is an adaptation of the TRIPOD approach geared toward a radiomics-specific context. The tool has been used throughout the literature for evaluating the methodological quality of radiomics studies, including applications to medical imaging [38]. Radiomics-based approaches for interpreting medical images have evolved to encompass the AI techniques of classical ML and, most recently, deep learning models. Most recently, in recognition of the growing need for an evaluation tool specific to AI applications in medical imaging, the Checklist for AI in Medical Imaging (CLAIM) was published in 2020 [39]. The 42 elements of CLAIM aim to be a best practice guide for authors presenting their research on applications of AI in medical imaging, ranging from classification and image reconstruction to text analysis and workflow optimization. Other tools—the Quality Assessment of Diagnostic Accuracy Studies (QUADAS-2) tool [40] and Prediction model Risk Of Bias ASsessment Tool (PROBAST) [41]—importantly evaluate the risk of bias in studies based on what is reported about their models. Bias relates to systematic limitations or flaws in study design, methods, execution, or analysis that distort estimates of model performance [40]. High risk of bias discourages adaptation of the reported model outside of its original research context, and, at a systemic level, undermines model reproducibility and translation into clinical practice.
3.4.2. Reporting Quality and Risk of Bias in Image-Based Glioma Grade Prediction
3.4.3. Future of Reporting Guidelines and Risk of Bias Tools for ML Studies
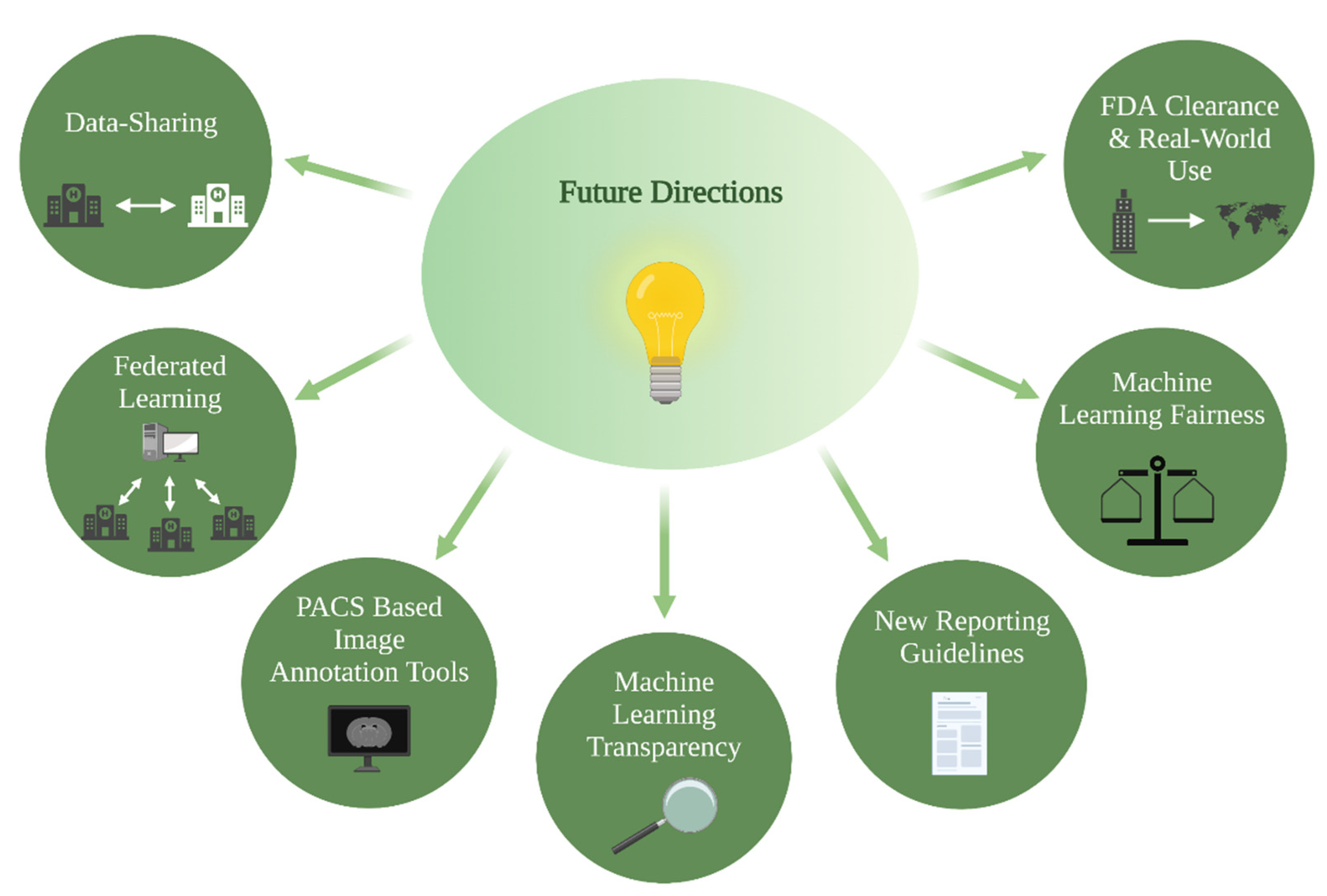
4. Future Directions
This entry is adapted from the peer-reviewed paper 10.3390/cancers14112623
References
- Yeo, M.; Kok, H.K.; Kutaiba, N.; Maingard, J.; Thijs, V.; Tahayori, B.; Russell, J.; Jhamb, A.; Chandra, R.V.; Brooks, M.; et al. Artificial intelligence in clinical decision support and outcome prediction–Applications in stroke. J. Med. Imaging Radiat. Oncol. 2021, 65, 518–528.
- Kaka, H.; Zhang, E.; Khan, N. Artificial Intelligence and Deep Learning in Neuroradiology: Exploring the New Frontier. Can. Assoc. Radiol. J. 2021, 72, 35–44.
- Sidey-Gibbons, J.A.M.; Sidey-Gibbons, C.J. Machine learning in medicine: A practical introduction. BMC Med. Res. Methodol. 2019, 19, 64.
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577.
- Aerts, H.J.W.L.; Velazquez, E.R.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006.
- Giger, M.L. Machine Learning in Medical Imaging. J. Am. Coll. Radiol. 2018, 15, 512–520.
- Ostrom, Q.T.; Cioffi, G.; Gittleman, H.; Patil, N.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2012–2016. Neuro-Oncology 2019, 21 Suppl. S5, v1–v100.
- Abdel Razek, A.A.K.; Alksas, A.; Shehata, M.; AbdelKhalek, A.; Abdel Baky, K.; El-Baz, A.; Helmy, E. Clinical applications of artificial intelligence and radiomics in neuro-oncology imaging. Insights Imaging 2021, 12, 152.
- Thon, N.; Tonn, J.-C.; Kreth, F.-W. The surgical perspective in precision treatment of diffuse gliomas. OncoTargets Ther. 2019, 12, 1497–1508.
- Hu, L.S.; Hawkins-Daarud, A.; Wang, L.; Li, J.; Swanson, K.R. Imaging of intratumoral heterogeneity in high-grade glioma. Cancer Lett. 2020, 477, 97–106.
- Seow, P.; Wong, J.H.D.; Annuar, A.A.; Mahajan, A.; Abdullah, N.A.; Ramli, N. Quantitative magnetic resonance imaging and radiogenomic biomarkers for glioma characterisation: A systematic review. Br. J. Radiol. 2018, 91, 20170930.
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.P.M.; Granton, P.; Zegers, C.M.L.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446.
- Buchlak, Q.D.; Esmaili, N.; Leveque, J.-C.; Bennett, C.; Farrokhi, F.; Piccardi, M. Machine learning applications to neuroimaging for glioma detection and classification: An artificial intelligence augmented systematic review. J. Clin. Neurosci. 2021, 89, 177–198.
- Chow, D.; Chang, P.; Weinberg, B.D.; Bota, D.A.; Grinband, J.; Filippi, C.G. Imaging Genetic Heterogeneity in Glioblastoma and Other Glial Tumors: Review of Current Methods and Future Directions. Am. J. Roentgenol. 2018, 210, 30–38.
- Pesapane, F.; Codari, M.; Sardanelli, F. Artificial intelligence in medical imaging: Threat or opportunity? Radiologists again at the forefront of innovation in medicine. Eur. Radiol. Exp. 2018, 2, 35.
- Pemberton, H.G.; Zaki, L.A.M.; Goodkin, O.; Das, R.K.; Steketee, R.M.E.; Barkhof, F.; Vernooij, M.W. Technical and clinical validation of commercial automated volumetric MRI tools for dementia diagnosis-a systematic review. Neuroradiology 2021, 63, 1773–1789.
- Richens, J.G.; Lee, C.M.; Johri, S. Improving the accuracy of medical diagnosis with causal machine learning. Nat. Commun. 2020, 11, 3923.
- Rubin, D.L. Artificial Intelligence in Imaging: The Radiologist’s Role. J. Am. Coll. Radiol. 2019, 16, 1309–1317.
- Brereton, R.G.; Lloyd, G.R. Support Vector Machines for classification and regression. Analyst 2010, 135, 230–267.
- Sohn, C.K.; Bisdas, S. Diagnostic Accuracy of Machine Learning-Based Radiomics in Grading Gliomas: Systematic Review and Meta-Analysis. Contrast Media Mol. Imaging 2020, 2020, 2127062.
- Gordillo, N.; Montseny, E.; Sobrevilla, P. State of the art survey on MRI brain tumor segmentation. Magn. Reson. Imaging 2013, 31, 1426–1438.
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444.
- Chartrand, G.; Cheng, P.M.; Vorontsov, E.; Drozdzal, M.; Turcotte, S.; Pal, C.J.; Kadoury, S.; Tang, A. Deep Learning: A Primer for Radiologists. RadioGraphics 2017, 37, 2113–2131.
- Ge, C.; Gu, I.Y.-H.; Jakola, A.S.; Yang, J. Deep Learning and Multi-Sensor Fusion for Glioma Classification Using Multistream 2D Convolutional Networks. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 17–22 July 2018; pp. 5894–5897.
- Kabir-Anaraki, A.; Ayati, M.; Kazemi, F. Magnetic resonance imaging-based brain tumor grades classification and grading via convolutional neural networks and genetic algorithms. Biocybern. Biomed. Eng. 2019, 39, 63–74.
- Ge, C.; Gu, I.Y.-H.; Jakola, A.S.; Yang, J. Deep semi-supervised learning for brain tumor classification. BMC Med. Imaging 2020, 20, 1–11.
- Sharif, M.I.; Li, J.P.; Khan, M.A.; Saleem, M.A. Active deep neural network features selection for segmentation and recognition of brain tumors using MRI images. Pattern Recognit. Lett. 2020, 129, 181–189.
- Hayashi, Y. Toward the transparency of deep learning in radiological imaging: Beyond quantitative to qualitative artificial intelligence. J. Med. Artif. Intell. 2019, 2, 19.
- Topol, E.J. High-performance medicine: The convergence of human and artificial intelligence. Nat. Med. 2019, 25, 44–56.
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans. Med. Imaging 2015, 34, 1993–2024.
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Prior, F. The Cancer Imaging Archive (TCIA): Maintaining and operating a public information repository. J. Digit. Imaging 2013, 26, 1045–1057.
- Madhavan, S.; Zenklusen, J.-C.; Kotliarov, Y.; Sahni, H.; Fine, H.A.; Buetow, K. Rembrandt: Helping Personalized Medicine Become a Reality through Integrative Translational Research. Mol. Cancer Res. 2009, 7, 157–167.
- Rajan, P.V.; Karnuta, J.M.; Haeberle, H.S.; Spitzer, A.I.; Schaffer, J.L.; Ramkumar, P.N. Response to letter to the editor on “Significance of external validation in clinical machine learning: Let loose too early?”. Spine J. 2020, 20, 1161–1162.
- Wesseling, P.; Capper, D. WHO 2016 Classification of gliomas. Neuropathol. Appl. Neurobiol. 2018, 44, 139–150.
- Brat, D.J.; Aldape, K.; Colman, H.; Figrarella-Branger, D.; Fuller, G.N.; Giannini, C.; Weller, M. cIMPACT-NOW update 5: Recommended grading criteria and terminologies for IDH-mutant astrocytomas. Acta Neuropathol. 2020, 139, 603–608.
- Collins, G.S.; Reitsma, J.B.; Altman, D.G.; Moons, K.G. Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis (TRIPOD): The TRIPOD Statement. Br. J. Surg. 2015, 102, 148–158.
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; van Timmeren, J.; Sanduleanu, S.; Larue, R.T.H.M.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762.
- Park, J.E.; Kim, H.S.; Kim, D.; Park, S.Y.; Kim, J.Y.; Cho, S.J.; Kim, J.H. A systematic review reporting quality of radiomics research in neuro-oncology: Toward clinical utility and quality improvement using high-dimensional imaging features. BMC Cancer 2020, 20, 29.
- Mongan, J.; Moy, L.; Kahn, C.E. Checklist for Artificial Intelligence in Medical Imaging (CLAIM): A Guide for Authors and Reviewers. Radiol. Artif. Intell. 2020, 2, e200029.
- Whiting, P.F.; Rutjes, A.W.S.; Westwood, M.E.; Mallett, S.; Deeks, J.J.; Reitsma, J.B.; Leeflang, M.M.; Sterne, J.A.; Bossuyt, P.M.; QUADAS-2 Group. QUADAS-2: A Revised Tool for the Quality Assessment of Diagnostic Accuracy Studies. Ann. Intern. Med. 2011, 155, 529–536.
- Wolff, R.F.; Moons, K.G.; Riley, R.D.; Whiting, P.F.; Westwood, M.; Collins, G.S. PROBAST: A Tool to Assess the Risk of Bias and Applicability of Prediction Model Studies. Ann. Intern. Med. 2019, 170, 51–58.
- Andaur Navarro, C.L.; Damen, J.A.A.; Takada, T.; Nijman, S.W.J.; Dhiman, P.; Ma, J.; Collins, G.S.; Bajpai, R.; Riley, R.D.; Moons, K.G.M.; et al. Risk of bias in studies on prediction models developed using supervised machine learning techniques: Systematic review. BMJ 2021, 20, n2281.
- Yao, A.D.; Cheng, D.L.; Pan, I.; Kitamura, F. Deep Learning in Neuroradiology: A Systematic Review of Current Algorithms and Approaches for the New Wave of Imaging Technology. Radiol. Artif. Intell. 2020, 2, e190026.
- Olivier, M.; Asmis, R.; Hawkins, G.A.; Howard, T.D.; Cox, L.A. The Need for Multi-Omics Biomarker Signatures in Precision Medicine. Int. J. Mol. Sci. 2019, 20, 4781.
- Subramanian, I.; Verma, S.; Kumar, S.; Jere, A.; Anamika, K. Multi-omics Data Integration, Interpretation, and Its Application. Bioinform. Biol. Insights 2020, 14, 1177932219899051.
