Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Mycoplankton are saprophytic organisms in plankton communities in marine and freshwater ecosystems. They consist of filamentous free-living fungi and yeasts associated with planktonic particles or phytoplankton. Similar to planktonic bacteria, these aquatic fungi play important roles in heterotrophic mineralization and nutrient cycling. Planktonic bacteria can be up to 20 mm in diameter and over 50 mm in length
- Mycoplankton
- Ecological Roles
- Cycling
1. Abundance of Mycoplankton
A typical milliliter of seawater is known to contain about 1000 fungal cells [1]. The abundance of fungi has been often estimated by researchers using culturable, microscopic, or molecular methods. However, due to ‘great plate anomaly’ and other biases, the densities of culturable fungi in the ocean are several orders of magnitude lower than that of fungi detected either by direct detection or molecular techniques. The culturable fungal abundance (CFU L−1) was found to be three orders of magnitude [2][3], while the abundance (gene copies L−1) based on the qPCR method was five to eight orders of magnitude [4][5][6][7]. Fungal enumeration by culturing has been criticized because a colony can arise out of single spores, groups of spores, single cells, or mycelial fragments. Therefore, methods based on direct detection of fungal hyphae or ergosterol and qPCR have been developed (Table 1). Even though these alternative methods have their own biases, they are much less time-consuming and labor-intensive and provide reasonably reliable estimates of fungal abundance.
Table 1. Abundance of planktonic fungi in various oceanic regions estimated by different methods and their comparison with bacterial abundance.
| Estimation Method for Fungi | Sampling Region |
Fungal Abundance |
Bacterial Abundance | Reference |
|---|---|---|---|---|
| Biomass carbon | Coastal Chile | 0.03–6 μg C L−1 | - | [8] |
| Biomass carbon | Coastal Chile | 0.01–40 μg C L−1 | 10–44 μg C L−1 | [9] |
| Fatty Acid (18:2ω6) | Coastal Chile | 0.1–3 μg L−1 | 10–44 μg C L−1 | [9] |
| Ergosterol | Arctic waters | 1.02 μg C L−1 | 5 to >25 μg C L−1 | [10][11] |
| qPCR (DNA concentration) | West Pacific Warm Pool | Basidiomycota (max. 10 ng μL−1, open-ocean station) Ascomycota (max. 14 pg μL−1, coastal station) | ~10 ng μL−1 | [12] |
| qPCR (18S rRNA gene copy number) | Coastal Plymouth, Western English Channel | 5.1 × 105 to 9.9 × 107 copies L−1 | 0.2 × 106–1.6 × 106 cells mL−1 | [5][13] |
| qPCR (18S rRNA gene copy number) | Coastal region, Bohai Sea | 4.28 × 106 to 1.13 × 107 copies L−1 | ~ 2 × 106 cells L−1 | [4] |
| qPCR (18S rRNA gene copy number) | PICO | 1.0 × 107 to 7.54 × 108 copies L−1 |
- | [14] |
“-” = data not available.
Fungal filaments, ranging from 1–3 μm in diameter and 10–200 μm in length were detected as individual filaments or aggregates in the coastal upwelling ecosystem off the coast of Central Chile using the Calcofluor White staining method. This aggregate formation was associated with the efficient remineralization of organic matter in seawater [8]. The vertical profile of fungal biomass showed higher values at the surface compared to greater depths and agreed with those of phytoplankton biomass and physicochemical parameters, suggesting higher fungal activity during high organic matter availability in a coastal upwelling ecosystem off the coast of Chile [9]. In the same study, the fungal biomass determined by the abundance of hyphae positively correlated with phospholipid fatty acid (18:2ω6), a fungal biomarker, and reflected the degradation of protein and carbohydrate polymers. Of interest, the fungal biomass (0.04 μgCL−1 to 40 μgCL−1) was comparable to prokaryotic biomass (10 μgCL−1 to 44 μgCL−1) and both biomasses peaked upon a decline in phytoplankton biomass, suggesting that the availability of detritus determined their abundances. Such an association of fungal abundance was also evident from studies that were based on molecular techniques [5][14].
The analysis of the abundances of major planktonic fungi (Ascomycota and Basidiomycota) in a transect from the Hawaiian coast to Australia revealed that Ascomycota had a high abundance only in coastal stations, whereas Basidiomycota was high in both oceanic and coastal stations [12]. The abundance of Basidiomycota (maximum 10 ng/μL, open-ocean station) was much higher than that of Ascomycota (maximum 14 pg/μL, coastal station) and similar to that of bacterioplankton in all the stations. The abundance of mycoplankton was highest at the surface, a pattern similar to that exhibited by bacterioplankton in most stations. In a high-resolution time-series study at PICO, an abundance of up to eight orders of magnitude was observed with two peaks each year, one each in summer and fall. The abundance was found to exhibit a dynamic pattern and was linked to chlorophyll a, SiO4, and oxygen saturation. As PICO is a site with a high salinity, no correlation was observed between abundance and salinity [14]. Conversely, mycoplankton abundance was shown to positively correlate with particulate organic carbon, ammonia, total particulate nitrogen, and particulate organic nitrogen, while negatively with salinity at the coastal Plymouth site [5]. The negative correlation with salinity was attributed to an increased abundance due to riverine inputs. Whereas the factors with a positive correlation were the growth substrates that increase with autochthonous production or allochthonous inputs [5]. Sites that experience river inflows are generally reported to contain fungal and nutritional inputs from terrestrial sources [8][15][16].
Taken together, these studies reveal the ubiquitous presence and high abundance of mycoplankton within marine environments. Evidence of mycoplankton abundance similar to that of bacterioplankton in nutrient-rich habitats emphasizes that mycoplankton are an important component of coastal realms. The association of mycoplankton with environmental factors suggests their important role in detrital processing and nutrient cycling. The paucity of knowledge on mycoplankton abundance patterns in the pelagic realm warrants future investigations.
2. Ecological Roles of Mycoplankton
Fungi in the transition zones of salt marshes and mangroves were found to play the roles of saprobes, symbionts, pathogens, and parasites, similar to their terrestrial counterparts [17]. Currently, their roles are yet to be established, especially in the open-ocean water column, even though they have been detected in the entire marine water column. Arguably, a reliance on osmotrophy determines the ecology of fungi in marine ecosystems similar to terrestrial, and therefore nutrient-rich environments have both abundant and diverse fungi. In addition, the ability to attach to detritus or particulate organic matter enables fungi to grow in the flowing and turbulent water of the oceans. Thus, owing to these two important aspects of fungal feeding strategies (i.e., osmotrophy and attachment to the substrate), marine fungi are generally considered to play the roles of decomposers, parasites, and denitrifiers [18][19]. Unfortunately, only a few studies provide direct evidence of their ecological roles; thus, fungi are often neglected in the ocean ecosystem models [12][20]. Nevertheless, with the piling evidence of their contribution to the marine ecosystems, marine microbiologists have started to realize their importance in nutrient cycling and the food web. For example, laboratory-based physiological studies [21], biomass [22], direct detection of fungal mycelia [9][23], zoospores and rhizoid structures on host cells [24][25], metabolic potential/physiological diversity analysis [26], the high copy number of rRNA gene [5][7][12], live fungal biomass (ergosterol) [10][27] provide indications of their viability and possible ecological roles in the water column. The following sub-sections discuss the predicted ecological roles of fungi in marine environments, which are illustrated in Figure 1.
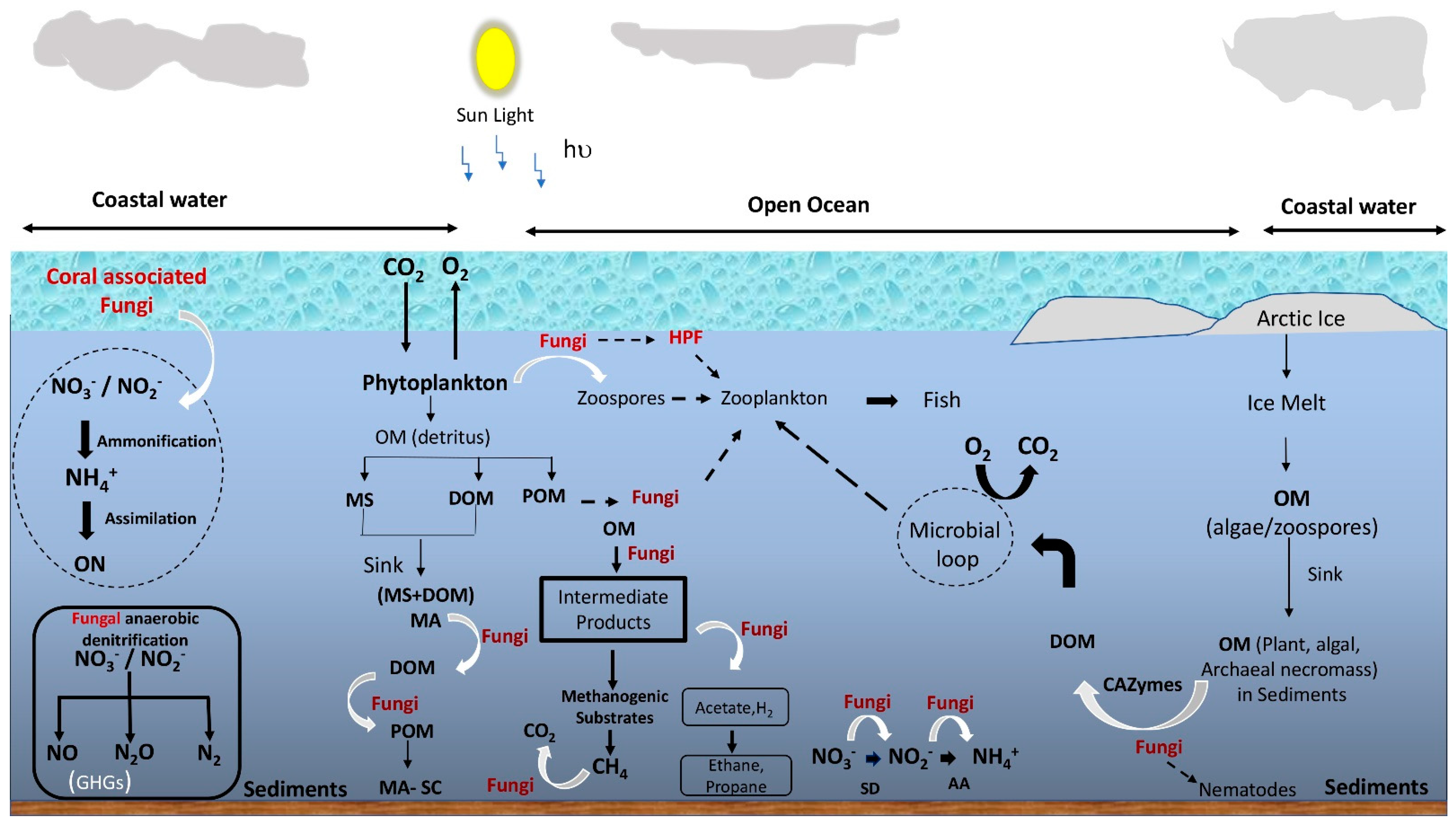
Figure 1. Schematic representation of the overview of possible roles of fungi in the marine food web and biogeochemical cycling. OM = organic matter; DOM = dissolved organic matter; MS = marine snow; POM = particulate organic matter; HPF = hyper parasitic fungi; SD = suboxic denitrification; AA = anaerobic ammonification; GHG = greenhouse gas; MA = macroaggregate; MA-SC = macroaggregate-sequestered carbon; ON = organic nitrogen. Black dotted arrows indicate feeding and white curved arrows indicate fungal involvement in the conversion.
2.1. Biogeochemical Cycling
2.1.1. Role in Organic Matter Decomposition and Aggregation
Marine ecosystems receive and process a large amount of bio-recalcitrant, terrigenous organic matter (particulate) often in the form of lignocellulosic substrates. In addition, a large pool of bio-labile organic matter (dissolved and particulate) in the ocean is produced from algal detritus. These forms of organic matter in the ocean are mainly recycled by microbial decomposers such as bacteria and fungi. Compared to bacteria, fungi can more efficiently mineralize lignocellulosic substrates due to their lower metabolic nutrient demand and wider enzymatic capabilities [28]. The decomposer role of fungi in aquatic ecosystems is mainly known from lotic systems, mangroves, and wetlands [29][30][31]. However, the frequent isolation of marine fungi from floating, sunken woody substrates, and plant detritus [32][33] also suggests such a role in coastal and pelagic ecosystems.
The colonization of lignocellulosic substrates by marine fungi is extensively studied [34]. However, there are fewer studies on their ability to utilize the lignocellulosic materials in the environment. In vitro studies suggest that marine fungi have the potential to degrade lignocellulosic components by their ability to produce hydrolytic enzymes, such as laccase, cellulase, amylase, alginase, laminarinase, peroxidase, pectinase, and xylanase [35][36][37][38][39]. Marine ascomycetes and basidiomycetes were demonstrated to solubilize significant amounts of lignin from wood in vitro, suggesting that they can carry out a ‘white-rot like’ role in the marine environment [40][41][42][43]. For example, the basidiomycetes Nia vibrissa, which were isolated from wood submerged in the sea, caused a pattern of wood decay characteristic of the white-rot type when cultured on different wood species [41]. Historically, the morphological decay features observed in woody biomass colonized by marine fungi were indicative of soft-rot and white-rot decay [44]. The soft-rot strategy is efficient in marine systems compared to white rot, which suffers from the leaching of lignocellulolytic enzymes into the surroundings [34][37][45][46]. Typically, the soft-rot strategy involves extensive cellulose and hemicellulose degradation with limited lignin degradation, and such a strategy is key to the survival of ascomycetes in oceanic waters [46]. The prevalence of the soft-rot strategy in oceanic waters was evident from the dominance of ascomycetes and disappearance of basidiomycetes with the prolonged submersion of the woody substrate (driftwood) in the Arctic Ocean [46]. The above findings perhaps advocate that these marine fungi, armored with lignocellulolytic activity, are most likely capable of degrading lignocellulosic substrates in both coastal and oceanic waters by colonization.
Apart from lignocellulosic substrates, marine fungi are also capable of processing algal polymeric substrates by secreting a plethora of hydrolytic enzymes in vitro [38]. It has been shown that the hydrolytic activity of fungi increases in presence of phytoplankton-derived biopolymers, and such activity can process about 30% photosynthetic carbon in a coastal upwelling system [9]. Moreover, in the coastal water column, fungi are found to grow during productive periods of high substrate availability and feature high hydrolytic activity [9]. Later experiments demonstrated the assimilation of 13C-labeled algal transparent exopolysaccharides (TEP) and the accumulation of 13C in Cladosporium (Ascomycota) and Malassezia (Basidiomycota), which provide direct evidence for the utilization of algal polysaccharides by saprotrophic planktonic fungi [47].
Some studies show that fungi in marine environments produce macroaggregates from DOM without the need for nucleation, where the presence of fungal hyphae makes the macroaggregates stable and renders them less easily degradable [22]. Such evidence of macroaggregates in deep-sea regions is predicted to lead to long-term carbon sequestration, ultimately affecting the carbon biogeochemical cycling and global weather change [22]. A similar aggregate formation was also observed in the coastal water column, and it was suggested that the combined action of fungi and bacteria could result in a highly efficient microbioreactor able to process particulate organic matter (POM) and DOM during sedimentation [8]. Furthermore, it is speculated that fungi contribute to organic matter degradation in the deep sea owing to their dominance in the overall biomass within marine aggregates (snow) [48]. These findings suggest that planktonic fungi play a role in the formation and stabilization of the marine aggregates and their simultaneous degradation to DOM. Interestingly, such a contribution highlights their possible link to the POM-DOM cycling in the ocean (Figure 1). Thus, fungi might play a much more important role in biological carbon pumps or ocean carbon storage than what is currently perceived.
As fungi are known to produce a variety of enzymes that have the potential to break down the chemical bonds of plastic polymers, they might have a role in the degradation of marine plastics [49]. Seminal works on plastic deterioration by marine fungi have suggested that polyurethanes are more susceptible to fungal attacks [50]. Interestingly, a recent study reported that fungi (e.g., Aspergillus flavus, A. terreus, A. niger, A. fumigatus, and Penicillium sp.) isolated from seawater are potential degraders of polyethylene [51]. These reports highlight the underestimated role of planktonic fungi as degraders of marine plastic wastes.
Considering earlier studies, it is not surprising that marine fungi can carry out the role of saprotrophs in the coastal and ocean waters. With their unique ability to produce a myriad of hydrolytic enzymes and the colonization of lignocellulosic substrates, algal biopolymers, marine snow, and plastics, marine fungi might contribute to the process of microbial carbon sequestration in the ocean. Their role in long-term carbon sequestration, however, remains speculative and needs further investigation.
2.1.2. Role in Nutrient Metabolism
With the recent application of omics and microarray techniques, people's ability to understand the mechanisms underpinning the function of marine fungi in biogeochemical cycling is accelerating. Particularly, studies based on metagenomics and metatranscriptomics provided evidence for the fungi-associated metabolic processes in the marine and water columns. Metagenomic studies discovered genes involved in amino acid metabolism, the aerobic carboxylation of glucose, anaerobic decarboxylation of pyruvate, urea, sulfur metabolism, etc. [52][53]. Fungal genes involved in complex C and fatty acid metabolism have been found across all depths and regions, and it is suggested that fungi might replace phytoplankton for vitamin supplies in deep waters [53]. Similarly, metatranscriptomics also revealed fungal transcripts that were assigned to protein, carbohydrate, and lipid metabolism [54]. Some studies based on metatranscriptomics prove the presence of only fungal carbohydrate-active enzymes (CAZymes) and carbohydrate-binding modules in the secreted proteome, suggesting active carbohydrate (microbial cell envelopes, plant, and algal detritus) degradation by fungi and their involvement in carbon cycling in the ecosystems [55]. Metagenome prediction using the PICRUSt2 tool suggests that fungal communities in marine waters are primarily aerobic and acquire energy through the oxidation of fatty acids [56]. This study also suggested that the metabolism of amino acids, carbohydrates and energy, fatty acids and lipids, nitrogen, sulfur, and other compounds, such as vitamins, octane, methyl ketone, heme, and secondary metabolite, possibly represent the core metabolism of marine mycoplankton in marine habitats ranging from estuarine to open ocean. In addition, high CAZymes per gene suggested that pelagic fungi are active in carbohydrate degradation [57].
A few earlier studies have shown the presence of fungi of known and new taxonomic groups in methane hydrates [58], suggesting their possible role in carbon flux fueled by methane, similar to the methanotrophic prokaryotes [59]. Although, methane-utilizing yeasts were reported much earlier [60], a more recent study revealed the significant correlation of the members of marine yeast, Cryptococcus curvatus, with methane and ethane [61]. This suggests the involvement of fungi in methane cycling in the ocean and their probable interactions with methanogenic or ethanogenic prokaryotes. In addition, fungi are proposed to be H2 producers that help in the growth and survival of sulfate-reducing bacteria in the deep ocean, indicating their possible involvement in the anaerobic oxidation of methane [62]. A few studies also suggested the role of fungi in nitrogen cycling in the ocean. Marine fungi were found to be associated with nitrate reduction, nitrite accumulation, and ammonia formation in the anoxic region of the ocean [21], denitrification, co-denitrification, ammonification [63], and nitrite reduction in the deep biosphere [54]. Using GeoChip, several fungal genes were detected that catalyze ammonification from nitrite and urea, ammonia assimilation, and denitrification in marine sediment [10]. Another line of evidence suggests that endolithic fungi were involved in at least two processes of the nitrogen cycle within corals: (1) reduction of nitrate and/or nitrite to ammonia, and (2) ammonia assimilation for biosynthesis [64]. As marine fungi are capable of anaerobic denitrification with the formation of greenhouse gases (NO and N2O) and nitrogen (N2) [65], their impact on global climate should be further explored.
Overall, these findings suggest fungi as an important component of nutrient cycling (both carbon and nitrogen) in the ocean and warrant their inclusion in marine microbial ecosystem models involving biogeochemical cycling.
2.2. Fungal Contribution to the Marine Food Web and Biotic Interactions
Mycoplankton are known to play an important role within the marine microbial food web as diatom parasites [5][24][66]. Particularly, chytrids found in the coastal water column, open ocean, and Arctic regions are reported to channel organic matter and energy to higher trophic levels converting inedible phytoplankton to zoospores (high in polyunsaturated fatty acids and cholesterol) that serve as food for zooplankton [25][67][68]. This mechanism, known as the mycoloop, provides nutrients to the food web through the zoospores of either parasitic fungi or saprotrophic fungi. Fungi feed on substrates inedible for zooplankton, and in turn, produce zoospores rich in nutrients that are palatable to zooplankton [31]. These zoospores become a major food source, especially when inedible food sources predominate, thus making fungi responsible for the growth and reproduction of zooplankton [47][69]. The non-grazed zoospores, in turn, might contribute to the DOM and the detritus pool [68]. In addition, the fungi that are diatom parasites might prove to be successful competitors against zooplankton by controlling energy flow and food web dynamics [70]. Thus, the fungus–zooplankton association may alter the food web dynamics by either increasing the population of zooplankton or decreasing it. Furthermore, fungi may also serve as hosts for hyper-parasites, thereby reducing the parasitic load on the phytoplankton, and owing to their smaller size, hyperparasites, in turn, are grazed by zooplankton [71]. A tripartite interaction between Cryptomycota (hyperparasite), Chytridiomycota (parasite, saprotroph), and phytoplankton [4], and the niche separation between Cryptomycota (algal parasite) and Chytridiomycota have been speculated [72]. A recent study also revealed that Rozellomycota fungi, which are dominant during pre- and early bloom stages, have the potential to fuel a marine mycoloop [73]. Direct evidence of fungal parasitism in the marine water column, especially in the oceanic water column, is scarce, and thus would be an interesting topic of further exploration.
The organic detritus and its associated microbes are important to the marine food web. Fungi can convert the detritus into palatable forms for detritivores owing to their lignocellulose degradation capability, and are thus suggested to play an important role in the coastal water column and/or open-ocean detrital dynamics [9][32]. Fungi and bacteria occupy different functional niches in the decomposition of POM, wherein fungi act as primary degraders of particulate, and bacteria act as rapid recyclers of nutrient-rich organic matter compounds (e.g., algal biopolymers) [30]. Towards the decaying stages of a diatomic bloom, diatom secretes a large amount of mucus that forms aggregates (marine snow) in the water column, which are likely to act as chemical cues for colonization by fungal zoospores. These aggregates contribute to pelagic detritus, and upon sedimentation, they are transported along with the attached fungi to the deep sea [9]. Similarly, the colonization of transparent, exopolymeric particles (TEPs) by fungal hyphae was also observed, suggesting the possible transportation of the fungal mycelia-bound TEPs to the ocean’s sediment [23]. Marine snow or TEP-associated fungi possibly re-mineralize the polysaccharides therein and contribute to the bulk of DOM in the deep sea [32]. As marine snow aggregates are recalcitrant to bacterial degradation, the saprotrophic action of fungi supports bacterial metabolism by making DOM available to bacteria. Another piece of evidence for the fungal utilization of algal TEPs suggests the possible interactions (e.g., competition and syntropy) between bacteria and planktonic fungi [47][74]. Although fungi and bacteria were found to co-exist in the water column and serve as food sources for zooplankton, the role of fungi might be more significant than bacteria as they prevent decoupling between primary and secondary production and transfer carbon up the marine food web [75].
Marine fungi were acknowledged for their great importance under ocean acidification [3]. In fact, under ocean acidification, a reduction in Chytridiomycetes and Cryptomycota was reported [76]. It was further suggested that a decrease in the number of these parasitic members might lead to a subsequent increase in large phytoplankton and a decrease in small phytoplankton. This would alter the food web structure and may lead to a decrease in zooplankton. Moreover, an increase in the abundance of pathogenic fungi was also observed with acidification [76]. Several lines of evidence gathered from culturable and molecular studies of water columns suggest the presence of fungi that are known pathogens of plants, vertebrates, and invertebrates [76][77][78][79]. Thus, there might be an increase in pathogenic fungal abundance with progress in ocean acidification, leading to the breakdown of ecosystems. Planktonic fungi are also susceptible to viral infection, and because viruses are abundant (107–108 particles/mL) in marine environments, their lysis of planktonic fungi might also contribute to food web dynamics [80]. A few studies report the presence of mycoviruses in marine ecosystems and that the viral lysis of fungi might contribute to another pathway of carbon flow into the DOM pool [10][81]. However, further investigations are needed to clearly understand the interactions between fungi and viruses in the water column.
Marine fungi are the components of a complex matrix of multipartite interactions and play an important role in the food web as both saprotroph (consumer) and progenitor of zoospores (secondary producer). Further studies involving empirical dynamic modeling approaches, such as linear (multiple autoregressive models) and non-linear (convergent cross-mapping) models, can shed light on food web dynamics by generating data for a network analysis of such chaotic systems [80]. Currently, network analyses have mainly been used to understand the spatial distribution of marine fungal OTUs among sampling sites [18], sea regions and temperatures [82], and competitive and cooperative relationships within OTUs [4]. Additionally, network models could decipher the relationships within fungal taxa, and between fungi and other eukaryotes (primary producers, fungal predators) cohabiting a freshwater lake [83]. Nevertheless, the modeling approaches mentioned above can also contribute to the understanding of the host–parasite relationship and its consequences on the food web, estimation of energy and matter transfer in the food web, and co-occurrence of fungal groups and their correlation with physicochemical and biological variables [18][68][80][82].
References
- Kubanek, J.; Jensen, P.R.; Keifer, P.A.; Sullards, M.C.; Collins, D.O.; Fenical, W. Seaweed resistance to microbial attack: A targeted chemical defense against marine fungi. Proc. Natl. Acad. Sci. USA 2003, 100, 6916–6921.
- Velmurugan, S.; Prasannakumar, C.; Manokaran, S.; Ajith Kumar, T.; Samkamaleson, A.; Palavesam, A. DNA barcodes for marine fungal identification and discovery. Fungal Ecol. 2013, 6, 408–418.
- Krause, E.; Wichels, A.; Giménez, L.; Gerdts, G. Marine fungi may benefit from ocean acidification. Aquat. Microb. Ecol. 2013, 69, 59–67.
- Wang, Y.; Sen, B.; He, Y.; Xie, N.; Wang, G. Spatiotemporal Distribution and Assemblages of Planktonic Fungi in the Coastal Waters of the Bohai Sea. Front. Microbiol. 2018, 9, 584.
- Taylor, J.D.; Cunliffe, M. Multi-year assessment of coastal planktonic fungi reveals environmental drivers of diversity and abundance. ISME J. 2016, 10, 2118–2128.
- Xu, W.; Pang, K.L.; Luo, Z.H. High fungal diversity and abundance recovered in the deep-sea sediments of the Pacific Ocean. Microb. Ecol. 2014, 68, 688–698.
- Le Calvez, T.; Burgaud, G.; Mahe, S.; Barbier, G.; Vandenkoornhuyse, P. Fungal diversity in deep-sea hydrothermal ecosystems. Appl. Environ. Microbiol. 2009, 75, 6415–6421.
- Gutiérrez, M.H.; Pantoja, S.; Quiñones, R.A.; González, R.R. First record of filamentous fungi in the coastal upwelling ecosystem off central Chile. Gayana (Concepción) 2010, 74, 66–73.
- Gutiérrez, M.H.; Pantoja, S.; Tejos, E.; Quiñones, R.A. The role of fungi in processing marine organic matter in the upwelling ecosystem off Chile. Mar. Biol. 2011, 158, 205–219.
- Hassett, B.T.; Borrego, E.J.; Vonnahme, T.R.; Rämä, T.; Kolomiets, M.V.; Gradinger, R. Arctic marine fungi: Biomass, functional genes, and putative ecological roles. ISME J. 2019, 13, 1484–1496.
- Sherr, E.B.; Sherr, B.F.; Fessenden, L. Heterotrophic protists in the Central Arctic Ocean. Deep Sea Res. Part II Top. Stud. Oceanogr. 1997, 44, 1665–1682.
- Wang, X.; Singh, P.; Gao, Z.; Zhang, X.; Johnson, Z.I.; Wang, G.Y. Distribution and diversity of planktonic fungi in the West Pacific Warm Pool. PLoS ONE 2014, 9, e101523.
- Mary, I.; Cummings, D.; Biegala, I.; Burkill, P.; Archer, S.; Zubkov, M. Seasonal dynamics of bacterioplankton structure at a coastal station in the western English Channel. Aquat. Microb. Ecol. 2006, 42, 119–126.
- Duan, Y.; Xie, N.; Song, Z.; Ward, C.S.; Yung, C.-M.; Hunt, D.E.; Johnson, Z.I.; Wang, G. A High-resolution Time-series Reveals Seasonal Patterns of Planktonic Fungi at a Temperate Coastal Ocean Site (Beaufort, North Carolina, USA). Appl. Environ. Microbiol. 2018, 84, e00967-18.
- Burgaud, G.; Woehlke, S.; Rédou, V.; Orsi, W.; Beaudoin, D.; Barbier, G.; Biddle, J.F.; Edgcomb, V.P. Deciphering the presence and activity of fungal communities in marine sediments using a model estuarine system. Aquat. Microb. Ecol. 2013, 70, 45–62.
- Vargas-Gastélum, L.; Riquelme, M. The Mycobiota of the Deep Sea: What Omics Can Offer. Life 2020, 10, 292.
- Jones, E.B.G.; Pang, K.-L.; Abdel-Wahab, M.A.; Scholz, B.; Hyde, K.D.; Boekhout, T.; Ebel, R.; Rateb, M.E.; Henderson, L.; Sakayaroj, J.; et al. An online resource for marine fungi. Fungal Divers. 2019, 96, 347–433.
- Zhang, T.; Fei Wang, N.; Qin Zhang, Y.; Yu Liu, H.; Yan Yu, L. Diversity and distribution of fungal communities in the marine sediments of Kongsfjorden, Svalbard (High Arctic). Sci. Rep. 2015, 5, 14524.
- Richards, T.A.; Jones, M.D.; Leonard, G.; Bass, D. Marine fungi: Their ecology and molecular diversity. Ann. Rev. Mar. Sci. 2012, 4, 495–522.
- Worden, A.Z.; Follows, M.J.; Giovannoni, S.J.; Wilken, S.; Zimmerman, A.E.; Keeling, P.J. Rethinking the marine carbon cycle: Factoring in the multifarious lifestyles of microbes. Science 2015, 347, 1257594.
- Cathrine, S.J.; Raghukumar, C. Anaerobic denitrification in fungi from the coastal marine sediments off Goa, India. Mycol. Res. 2009, 113, 100–109.
- Damare, S.; Raghukumar, C. Fungi and macroaggregation in deep-sea sediments. Microb. Ecol. 2008, 56, 168–177.
- Damare, S.; Raghukumar, C.; Raghukumar, S. Fungi in deep-sea sediments of the Central Indian Basin. Deep Sea Res. Part I: Oceanogr. Res. Pap. 2006, 53, 14–27.
- Gutiérrez, M.H.; Jara, A.M.; Pantoja, S. Fungal parasites infect marine diatoms in the upwelling ecosystem of the Humboldt current system off central Chile. Environ. Microbiol. 2016, 18, 1646–1653.
- Hassett, B.T.; Gradinger, R. Chytrids dominate arctic marine fungal communities. Environ. Microbiol. 2016, 18, 2001–2009.
- Fuentes, M.E.; Quiñones, R.A. Carbon utilization profile of the filamentous fungal species Fusarium fujikuroi, Penicillium decumbens, and Sarocladium strictum isolated from marine coastal environments. Mycologia 2016, 108, 1069–1081.
- Wang, Y.; Sen, K.; He, Y.; Xie, Y.; Wang, G. Impact of environmental gradients on the abundance and diversity of planktonic fungi across coastal habitats of contrasting trophic status. Sci. Total Environ. 2019, 683, 822–833.
- Fabian, J.; Zlatanovic, S.; Mutz, M.; Premke, K. Fungal-bacterial dynamics and their contribution to terrigenous carbon turnover in relation to organic matter quality. ISME J. 2017, 11, 415–425.
- Gulis, V.; Kuehn, K.; Suberkropp, K. The role of fungi in carbon and nitrogen cycles in freshwater ecosystems. In Fungi in Biogeochemical Cycles; Gadd, G.M., Ed.; Cambridge University Press: New York, NY, USA, 2006; pp. 404–435.
- Gessner, M.O.; Gulis, V.; Kuehn, K.; Chauvet, E.; Suberkropp, K. Fungal Decomposers of Plant Litter in Aquatic Ecosystems. In Environmental and Microbial Relationships, 2nd ed.; Kubicek, C.P., Druzhinina, I.S., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; pp. 301–324.
- Grossart, H.-P.; Van den Wyngaert, S.; Kagami, M.; Wurzbacher, C.; Cunliffe, M.; Rojas-Jimenez, K. Fungi in aquatic ecosystems. Nat. Rev. Microbiol. 2019, 17, 339–354.
- Raghukumar, S. The Role of Fungi in Marine Detrital Processes. In Marine Microbiology: Facets and Opportunities; Ramaiah, N., Ed.; National Institute of Oceanography: Goa, India, 2004; pp. 91–101.
- Jones, E.B.G. Fifty years of marine mycology. Fungal Divers. 2011, 50, 73–112.
- Pointing, S.B.; Hyde, K.D. Lignocellulose-degrading marine fungi. Biofouling 2000, 15, 221–229.
- Kamei, I.; Daikoku, C.; Tsutsumi, Y.; Kondo, R. Saline-dependent regulation of manganese peroxidase genes in the hypersaline-tolerant white rot fungus Phlebia sp. strain MG-60. Appl. Environ. Microbiol. 2008, 74, 2709–2716.
- Bonugli-Santos, R.C.; Durrant, L.R.; da Silva, M.; Sette, L.D. Production of laccase, manganese peroxidase and lignin peroxidase by Brazilian marine-derived fungi. Enzym. Microb. Technol. 2010, 46, 32–37.
- Arfi, Y.; Chevret, D.; Henrissat, B.; Berrin, J.-G.; Levasseur, A.; Record, E. Characterization of salt-adapted secreted lignocellulolytic enzymes from the mangrove fungus Pestalotiopsis sp. Nat. Commun. 2013, 4, 1810.
- Wang, Y.; Barth, D.; Tamminen, A.; Wiebe, M.G. Growth of marine fungi on polymeric substrates. BMC Biotechnol. 2016, 16, 3.
- Mainardi, P.H.; Feitosa, V.A.; Brenelli de Paiva, L.B.; Bonugli-Santos, R.C.; Squina, F.M.; Pessoa, A., Jr.; Sette, L.D. Laccase production in bioreactor scale under saline condition by the marine-derived basidiomycete Peniophora sp. CBMAI 1063. Fungal Biol. 2018, 122, 302–309.
- Bucher, V.V.C.; Hyde, K.; Pointing, S.; Reddy, C.A.; Reddy, S. Production of wood decay enzymes, mass loss and lignin solubilization in wood by marine ascomycetes and their anamorphs. Fungal Divers. 2003, 15, 1–14.
- Leightley, L.E.; Eaton, R.A. Nia vibrissa—A marine white rot fungus. Trans. Br. Mycol. Soc. 1979, 73, 35–40.
- Leightely, L.E. Wood decay activities of marine fungi. Bot. Mar. 1980, 23, 387–395.
- Jones, E.B.G.; Choeyklin, R. Chapter 10 Ecology of marine and freshwater basidiomycetes. In British Mycological Society Symposia Series; Boddy, L., Frankland, J.C., van West, P., Eds.; Academic Press: Cambridge, MA, USA, 2008; Volume 28, pp. 301–324.
- Mouzouras, R. Soft rot decay of wood by marine microfungi. J. Inst. Wood Sci. 1989, 11, 193–201.
- Hyde, K.D.; Jones, E.B.G.; Leaño, E.; Pointing, S.B.; Poonyth, A.D.; Vrijmoed, L.L.P. Role of fungi in marine ecosystems. Biodivers. Conserv. 1998, 7, 1147–1161.
- Rämä, T.; Davey, M.L.; Nordén, J.; Halvorsen, R.; Blaalid, R.; Mathiassen, G.H.; Alsos, I.G.; Kauserud, H. Fungi Sailing the Arctic Ocean: Speciose Communities in North Atlantic Driftwood as Revealed by High-Throughput Amplicon Sequencing. Microb. Ecol. 2016, 72, 295–304.
- Cunliffe, M.; Hollingsworth, A.; Bain, C.; Sharma, V.; Taylor, J.D. Algal polysaccharide utilisation by saprotrophic planktonic marine fungi. Fungal Ecol. 2017, 30, 135–138.
- Bochdansky, A.B.; Clouse, M.A.; Herndl, G.J. Eukaryotic microbes, principally fungi and labyrinthulomycetes, dominate biomass on bathypelagic marine snow. ISME J. 2017, 11, 362–373.
- Zeghal, E.; Vaksmaa, A.; Vielfaure, H.; Boekhout, T.; Niemann, H. The Potential Role of Marine Fungi in Plastic Degradation–A Review. Front. Mar. Sci. 2021, 8, 738877.
- Jones, E.G.; Le Campion-Alsumard, T. The biodeterioration of polyurethane by marine fungi. Int. Biodeterior. Bull. 1970, 6, 119–124.
- Alshehrei, F. Biodegradation of Low Density Polyethylene by Fungi Isolated from Red Sea Water. Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 1703–1709.
- Mahé, S.; Rédou, V.; Calvez, T.L.; Vandenkoornhuyse, P.; Burgaud, G. Fungi in Deep-Sea Environments and Metagenomics. In The Ecological Genomics of Fungi; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2013; pp. 325–354.
- Morales, S.; Biswas, A.; Herndl, G.; Baltar, F. Global Structuring of Phylogenetic and Functional Diversity of Pelagic Fungi by Depth and Temperature. Front. Mar. Sci. 2019, 6, 131.
- Orsi, W.D.; Edgcomb, V.P.; Christman, G.D.; Biddle, J.F. Gene expression in the deep biosphere. Nature 2013, 499, 205–208.
- Orsi, W.D.; Richards, T.A.; Francis, W.R. Predicted microbial secretomes and their target substrates in marine sediment. Nat. Microbiol. 2018, 3, 32–37.
- Sen, K.; Bai, M.; Sen, B.; Wang, G. Disentangling the structure and function of mycoplankton communities in the context of marine environmental heterogeneity. Sci. Total Environ. 2021, 766, 142635.
- Baltar, F.; Zhao, Z.; Herndl, G.J. Potential and expression of carbohydrate utilization by marine fungi in the global ocean. Microbiome 2021, 9, 106.
- Lai, X.; Cao, L.; Tan, H.; Fang, S.; Huang, Y.; Zhou, S. Fungal communities from methane hydrate-bearing deep-sea marine sediments in South China Sea. ISME J. 2007, 1, 756–762.
- Ruff, S.E.; Biddle, J.F.; Teske, A.P.; Knittel, K.; Boetius, A.; Ramette, A. Global dispersion and local diversification of the methane seep microbiome. Proc. Natl. Acad. Sci. USA 2015, 112, 4015–4020.
- Wolf, H.J.; Hanson, R.S. Isolation and Characterization of Methane-utilizing Yeasts. Microbiology 1979, 114, 187–194.
- Rédou, V.; Ciobanu, M.C.; Pachiadaki, M.G.; Edgcomb, V.; Alain, K.; Barbier, G.; Burgaud, G. In-depth analyses of deep subsurface sediments using 454-pyrosequencing reveals a reservoir of buried fungal communities at record-breaking depths. FEMS Microbiol. Ecol. 2014, 90, 908–921.
- Drake, H.; Ivarsson, M.; Bengtson, S.; Heim, C.; Siljeström, S.; Whitehouse, M.J.; Broman, C.; Belivanova, V.; Åström, M.E. Anaerobic consortia of fungi and sulfate reducing bacteria in deep granite fractures. Nat. Commun. 2017, 8, 55.
- Mouton, M.; Postma, F.; Wilsenach, J.; Botha, A. Diversity and characterization of culturable fungi from marine sediment collected from St. Helena Bay, South Africa. Microb. Ecol. 2012, 64, 311–319.
- Wegley, L.; Edwards, R.; Rodriguez-Brito, B.; Liu, H.; Rohwer, F. Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ. Microbiol. 2007, 9, 2707–2719.
- Shoun, H.; Kim, D.-H.; Uchiyama, H.; Sugiyama, J. Denitrification by fungi. FEMS Microbiol. Lett. 1992, 94, 277–281.
- Li, Q.; Wang, X.; Liu, X.; Jiao, N.; Wang, G. Diversity of parasitic fungi associated with phytoplankton in Hawaiian waters. Mar. Biol. Res. 2016, 12, 294–303.
- Kagami, M.; Miki, T.; Takimoto, G. Mycoloop: Chytrids in aquatic food webs. Front. Microbiol. 2014, 5, 166.
- Grossart, H.-P.; Wurzbacher, C.; James, T.Y.; Kagami, M. Discovery of dark matter fungi in aquatic ecosystems demands a reappraisal of the phylogeny and ecology of zoosporic fungi. Fungal Ecol. 2016, 19, 28–38.
- Kagami, M.; de Bruin, A.; Ibelings, B.W.; Van Donk, E. Parasitic chytrids: Their effects on phytoplankton communities and food-web dynamics. Hydrobiologia 2007, 578, 113–129.
- Tillmann, U.; Hesse, K.-J.; Tillmann, A. Large-scale parasitic infection of diatoms in the Northfrisian Wadden Sea. J. Sea Res. 1999, 42, 255–261.
- Gleason, F.H.; Lilje, O.; Marano, A.V.; Sime-Ngando, T.; Sullivan, B.K.; Kirchmair, M.; Neuhauser, S. Ecological functions of zoosporic hyperparasites. Front. Microbiol. 2014, 5, 244.
- Wang, Y.; Guo, X.; Zheng, P.; Zou, S.; Li, G.; Gong, J. Distinct seasonality of chytrid-dominated benthic fungal communities in the neritic oceans (Bohai Sea and North Yellow Sea). Fungal Ecol. 2017, 30, 55–66.
- Priest, T.; Fuchs, B.; Amann, R.; Reich, M. Diversity and biomass dynamics of unicellular marine fungi during a spring phytoplankton bloom. Environ. Microbiol. 2021, 23, 448–463.
- Taylor, J.D.; Cunliffe, M. Coastal bacterioplankton community response to diatom-derived polysaccharide microgels. Environ. Microbiol. Rep. 2017, 9, 151–157.
- Agha, R.; Saebelfeld, M.; Manthey, C.; Rohrlack, T.; Wolinska, J. Chytrid parasitism facilitates trophic transfer between bloom-forming cyanobacteria and zooplankton (Daphnia). Sci. Rep. 2016, 6, 35039.
- Reich, M.; Wichels, A.; Panzer, K.; Krause, E.; Gimenez, L.; Gerdts, G. Impacts of a reduction in seawater pH mimicking ocean acidification on the structure and diversity of mycoplankton communities. Aquat. Microb. Ecol. 2017, 79, 221–223.
- Li, W.; Wang, M.; Burgaud, G.; Yu, H.; Cai, L. Fungal Community Composition and Potential Depth-Related Driving Factors Impacting Distribution Pattern and Trophic Modes from Epi- to Abyssopelagic Zones of the Western Pacific Ocean. Microb. Ecol. 2019, 78, 820–831.
- Gonçalves, V.N.; Vitoreli, G.A.; de Menezes, G.C.A.; Mendes, C.R.B.; Secchi, E.R.; Rosa, C.A.; Rosa, L.H. Taxonomy, phylogeny and ecology of cultivable fungi present in seawater gradients across the Northern Antarctica Peninsula. Extremophiles 2017, 21, 1005–1015.
- Sun, J.Y.; Song, Y.; Ma, Z.P.; Zhang, H.J.; Yang, Z.D.; Cai, Z.H.; Zhou, J. Fungal community dynamics during a marine dinoflagellate (Noctiluca scintillans) bloom. Mar. Environ. Res. 2017, 131, 183–194.
- Frenken, T.; Alacid, E.; Berger, S.A.; Bourne, E.C.; Gerphagnon, M.; Grossart, H.-P.; Gsell, A.S.; Ibelings, B.W.; Kagami, M.; Küpper, F.C.; et al. Integrating chytrid fungal parasites into plankton ecology: Research gaps and needs. Environ. Microbiol. 2017, 19, 3802–3822.
- Nerva, L.; Ciuffo, M.; Vallino, M.; Margaria, P.; Varese, G.C.; Gnavi, G.; Turina, M. Multiple approaches for the detection and characterization of viral and plasmid symbionts from a collection of marine fungi. Virus Res. 2016, 219, 22–38.
- Li, W.; Wang, M.M.; Wang, X.G.; Cheng, X.L.; Guo, J.J.; Bian, X.M.; Cai, L. Fungal communities in sediments of subtropical Chinese seas as estimated by DNA metabarcoding. Sci. Rep. 2016, 6, 26528.
- Rojas-Jimenez, K.; Wurzbacher, C.; Bourne, E.C.; Chiuchiolo, A.; Priscu, J.C.; Grossart, H.-P. Early diverging lineages within Cryptomycota and Chytridiomycota dominate the fungal communities in ice-covered lakes of the McMurdo Dry Valleys, Antarctica. Sci. Rep. 2017, 7, 15348.
This entry is offline, you can click here to edit this entry!
