The use of biomarkers in cancer diagnosis, therapy, and prognosis has been highly effective over several decades. Studies of biomarkers in cancer patients pre- and post-treatment and during cancer progression have helped identify cancer stem cells (CSCs) and their related microenvironments. These analyses are critical for the therapeutic application of drugs and the efficient targeting and prevention of cancer progression, as well as the investigation of the mechanism of the cancer development. Biomarkers that characterize CSCs have thus been identified and correlated to diagnosis, therapy, and prognosis. However, CSCs demonstrate elevated levels of plasticity, which alters their functional phenotype and appearance by interacting with their microenvironments, in response to chemotherapy and radiotherapeutics. In turn, these changes induce different metabolic adaptations of CSCs.
- biomarkers
- cancer progression
- cancer stem cells
- cell plasticity
- microenvironment
- reprogramming factors
- stem cell markers
1. Introduction
2. Perspective on the Therapeutic Use of Biomarkers
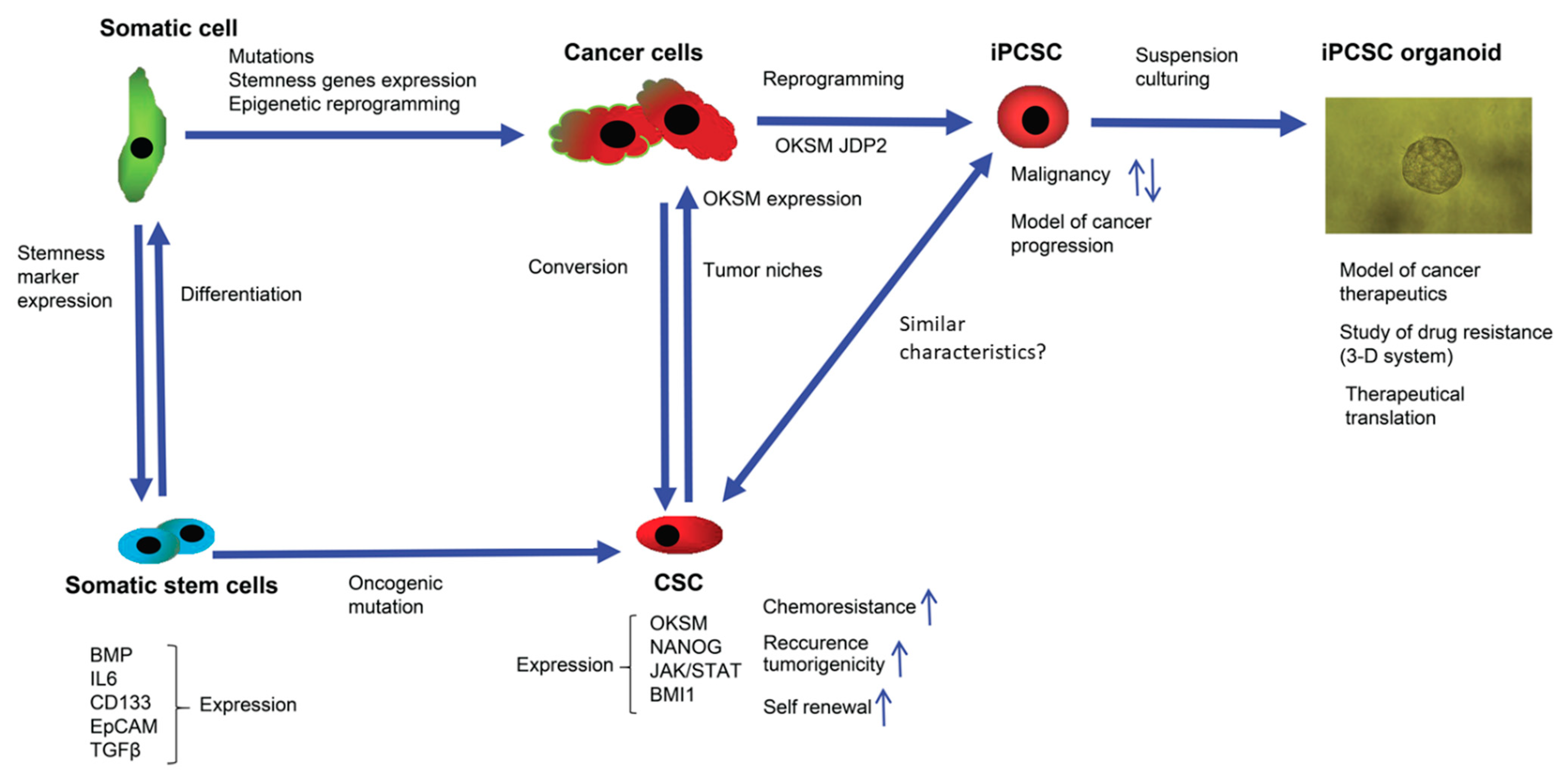
3. Elusive Problems Faced to Eliminate CSCs
4. Conclusions
This entry is adapted from the peer-reviewed paper 10.3390/jpm12050715
References
- Wuputra, K.; Ku, C.-C.; Wu, D.-C.; Lin, Y.-C.; Saito, S.; Yokoyama, K.K. Prevention of tumor risk associated with the reprogramming of human pluripotent stem cells. J. Exp. Clin. Cancer Res. 2020, 39, 100.
- Kreso, A.; Dick, J.E. Evolution of the cancer stem cell model. Cell Stem Cell 2014, 14, 275–291.
- Bonnet, D.; Dick, J.E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 1997, 3, 730–737.
- Reya, T.; Morrison, S.J.; Clarke, M.F.; Weissman, I.L. Stem cells, cancer, and cancer stem cells. Nature 2001, 414, 105–111.
- Singh, S.K.; Clarke, I.D.; Terasaki, M.; Bonn, V.E.; Hawkins, C.; Squire, J.; Dirks, P.B. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003, 63, 5821–5828.
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988.
- Pece, S.; Tosoni, D.; Confalonieri, S.; Mazzarol, G.; Vecchi, M.; Ronzoni, S.; Bernard, L.; Viale, G.; Pelicci, P.G.; Di Fiore, P.P. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell 2010, 140, 62–73.
- Schwitalla, S.; Fingerle, A.A.; Cammareri, P.; Nebelsiek, T.; Göktuna, S.I.; Ziegler, P.K.; Canli, O.; Heijmans, J.; Huels, D.J.; Moreaux, G.; et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell 2013, 152, 25–38.
- Ren, F.; Sheng, W.Q.; Du, X. CD133: A cancer stem cells marker, is used in colorectal cancers. World J. Gastroenterol. 2013, 19, 2603–2611.
- Zhang, S.; Cui, W. Sox2, a key factor in the regulation of pluripotency and neural differentiation. World J. Stem Cells 2014, 6, 305–311.
- Wuebben, E.L.; Rizzino, A. The dark side of SOX2: Cancer—a comprehensive overview. Oncotarget 2017, 8, 44917–44943.
- Yang, L.; Shi, P.; Zhao, G.; Xu, J.; Peng, W.; Zhang, J.; Zhang, G.; Wang, X.; Dong, Z.; Chen, F.; et al. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct. Target. Ther. 2020, 5, 8.
- Liu, B.; Yan, L.; Zhou, M. Target selection of CAR T cell therapy in accordance with the TME for solid tumors. Am. J. Cancer Res. 2019, 9, 228–241.
- Li, C.; Yan, Y.; Ji, W.; Bao, L.; Qian, H.; Chen, L.; Wu, M.; Chen, H.; Li, Z.; Su, C. OCT4 positively regulates Survivin expression to promote cancer cell proliferation and leads to poor prognosis in esophageal squamous cell carcinoma. PLoS ONE 2012, 7, e49693.
- Kuo, K.K.; Lee, K.T.; Chen, K.K.; Yang, Y.H.; Lin, Y.C.; Tsai, M.H.; Wuputra, K.; Lee, Y.L.; Ku, C.C.; Miyoshi, H.; et al. Positive Feedback Loop of OCT4 and c-JUN Expedites Cancer Stemness in Liver Cancer. Stem Cells 2016, 34, 2613–2624.
- Chiou, S.S.; Wang, S.S.; Wu, D.C.; Lin, Y.C.; Kao, L.P.; Kuo, K.K.; Wu, C.C.; Chai, C.Y.; Lin, C.L.; Lee, C.Y.; et al. Control of Oxidative Stress and Generation of Induced Pluripotent Stem Cell-like Cells by Jun Dimerization Protein 2. Cancers 2013, 5, 959–984.
- Saito, S.; Lin, Y.C.; Nakamura, Y.; Eckner, R.; Wuputra, K.; Kuo, K.K.; Lin, C.S.; Yokoyama, K.K. Potential application of cell reprogramming techniques for cancer research. Cell Mol. Life Sci. 2019, 76, 45–65.
- Miyoshi, N.; Ishii, H.; Nagai, K.; Hoshino, H.; Mimori, K.; Tanaka, F.; Nagano, H.; Sekimoto, M.; Doki, Y.; Mori, M. Defined factors induce reprogramming of gastrointestinal cancer cells. Proc. Natl. Acad. Sci. USA 2010, 107, 40–45.
- Walcher, L.; Kistenmacher, A.K.; Suo, H.; Kitte, R.; Dluczek, S.; Strauß, A.; Blaudszun, A.R.; Yevsa, T.; Fricke, S.; Kossatz-Boehlert, U. Cancer Stem Cells-Origins and Biomarkers: Perspectives for Targeted Personalized Therapies. Front. Immunol. 2020, 11, 1280.
- Kawamura, T.; Suzuki, J.; Wang, Y.V.; Menendez, S.; Morera, L.B.; Raya, A.; Wahl, G.M.; Izpisúa Belmonte, J.C. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 2009, 460, 1140–1144.
- Hong, H.; Takahashi, K.; Ichisaka, T.; Aoi, T.; Kanagawa, O.; Nakagawa, M.; Okita, K.; Yamanaka, S. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature 2009, 460, 1132–1135.
- Marión, R.M.; Strati, K.; Li, H.; Murga, M.; Blanco, R.; Ortega, S.; Fernandez-Capetillo, O.; Serrano, M.; Blasco, M.A. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature 2009, 460, 1149–1153.
- de Wet, L.; Williams, A.; Gillard, M.; Kregel, S.; Lamperis, S.; Gutgesell, L.C.; Vellky, J.E.; Brown, R.; Conger, K.; Paner, G.P.; et al. SOX2 mediates metabolic reprogramming of prostate cancer cells. Oncogene 2022, 41, 1190–1202.
- Metz, E.P.; Wuebben, E.L.; Wilder, P.J.; Cox, J.L.; Datta, K.; Coulter, D.; Rizzino, A. Tumor quiescence: Elevating SOX2 in diverse tumor cell types downregulates a broad spectrum of the cell cycle machinery and inhibits tumor growth. BMC Cancer 2020, 20, 941.
- Kwon, O.J.; Zhang, L.; Jia, D.; Zhou, Z.; Li, Z.; Haffner, M.; Lee, J.K.; True, L.; Morrissey, C.; Xin, L. De novo induction of lineage plasticity from human prostate luminal epithelial cells by activated AKT1 and c-Myc. Oncogene 2020, 39, 7142–7151.
- Saglam-Metiner, P.; Gulce-Iz, S.; Biray-Avci, C. Bioengineering-inspired three-dimensional culture systems: Organoids to create tumor microenvironment. Gene 2019, 686, 203–212.
- Varum, S.; Rodrigues, A.S.; Moura, M.B.; Momcilovic, O.; Easley, C.A.t.; Ramalho-Santos, J.; Van Houten, B.; Schatten, G. Energy metabolism in human pluripotent stem cells and their differentiated counterparts. PLoS ONE 2011, 6, e20914.
- Zong, W.X.; Rabinowitz, J.D.; White, E. Mitochondria and Cancer. Mol. Cell 2016, 61, 667–676.
- Yang, J.M.; Chi, W.Y.; Liang, J.; Takayanagi, S.; Iglesias, P.A.; Huang, C.H. Deciphering cell signaling networks with massively multiplexed biosensor barcoding. Cell 2021, 184, 6193–6206.e6114.
- Aramini, B.; Masciale, V.; Grisendi, G.; Bertolini, F.; Maur, M.; Guaitoli, G.; Chrystel, I.; Morandi, U.; Stella, F.; Dominici, M.; et al. Dissecting tumor growth: The role of cancer stem cells in drug resistance and recurrence. Cancers 2022, 14, 976.
- van der Pol, E.; Böing, A.N.; Harrison, P.; Sturk, A.; Nieuwland, R. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol. Rev. 2012, 64, 676–705.
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228.
- Balaj, L.; Lessard, R.; Dai, L.; Cho, Y.J.; Pomeroy, S.L.; Breakefield, X.O.; Skog, J. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat. Commun. 2011, 2, 180.
- Gaur, A.B.; Holbeck, S.L.; Colburn, N.H.; Israel, M.A. Downregulation of Pdcd4 by mir-21 facilitates glioblas toma proliferation in vivo. Neuro Oncol. 2011, 13, 580–590.
- Kim, M.S.; Haney, M.J.; Zhao, Y.; Mahajan, V.; Deygen, I.; Klyachko, N.L.; Inskoe, E.; Piroyan, A.; Sokolsky, M.; Okolie, O.; et al. Engineering macrophage-derived exosomes for targeted paclitaxel delivery to pulmonary metastases: In vitro and in vivo evaluations. Nanomedicine 2016, 12, 655–664.
- Bao, B.; Ahmad, A.; Azmi, A.S.; Ali, S.; Sarkar, F.H. Overview of cancer stem cells (CSCs) and mechanisms of their regulation: Implications for cancer therapy. Curr. Protoc. Pharmacol. 2013, 61, 14–25.
- Shultz, L.D.; Brehm, M.A.; Garcia-Martinez, J.V.; Greiner, D.L. Humanized mice for immune system investigation: Progress, promise and challenges. Nat. Rev. Immunol. 2012, 12, 786–798.
- Plaks, V.; Kong, N.; Werb, Z. The cancer stem cell niche: How essential is the niche in regulating stemness of tumor cells? Cell Stem Cell 2015, 16, 225–238.
- Sancho, P.; Barneda, D.; Heeschen, C. Hallmarks of cancer stem cell metabolism. Br. J. Cancer 2016, 114, 1305–1312.
- Du, F.Y.; Zhou, Q.F.; Sun, W.J.; Chen, G.L. Targeting cancer stem cells in drug discovery: Current state and future perspectives. World J. Stem Cells 2019, 11, 398–420.
