Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Oncology
Human T cell leukemia virus type 1 (HTLV-1) was identified as the first pathogenic human retrovirus and is estimated to infect 5 to 10 million individuals worldwide. Unlike other retroviruses, there is no effective therapy to prevent the onset of the most alarming diseases caused by HTLV-1, and the more severe cases manifest as the malignant phenotype of adult T cell leukemia (ATL). MicroRNA (miRNA) dysfunction is a common feature of leukemogenesis, and it is no different in ATL cases.
- HTLV-1
- T cell leukemia
- miRNAs
- carcinogenesis
1. HTLV-1 Viral Structure and Infection Mechanisms
HTLV-1 is the causative agent of ATL, a malignant tumor of CD4+ T cells. It is a retrovirus of the deltaretrovirus genus, and several subtypes of HTLV were discovered later—HTLV-2, HTLV-3, and HTLV-4. It has a structural organization like all retroviruses; two mature viral glycoproteins derived from a common precursor, an outer surface protein (SU) associated with a transmembrane (TM) protein, which is responsible for anchoring the SU-TM complex on the surface of the infected cell, an envelope consisting of a lipid bilayer of cellular origin, and the matrix protein located next to the membrane [1][2][3][4].
It is an enveloped double-stranded RNA virus, and its genome contains three structural genes: gag, pol, and env, and two regulatory genes, tax and rex. The tax and rex genes regulate the transactivation of viral replication, in addition to regulating the expression of viral proteins and the basic leucine zipper factor (HBZ) of HTLV-1, which are essential for the maintenance of viral persistence and pathogenesis, in addition to helping in the oncogenic process of ATL, stimulation of tumor growth and development [3][4][5][6].
Viral integration into the host’s DNA happens preferably at transcriptionally active regions, specifically near transcription factor binding sites. HTLV-1 uses different strategies to induce neoplastic transformation, but the immunogenic profile of the host is associated with inflammatory responses, promoting or protecting against the development of HAM or ATL. Its replication occurs parallel to the cell during mitosis, and its infection can occur vertically, during childbirth or breastfeeding, or parenterally, which consists of transfusions, transplants, intravenous drug use, and unprotected sexual intercourse [6][7][8][9][10][11].
Furthermore, HTLV infects mainly T lymphocytes but is also capable of infecting monocytes and dendritic cells due to its direct interaction with glucose transporter 1 (GLUT 1), which is ubiquitously expressed on cell surfaces (Figure 1) [11][12][13][14].
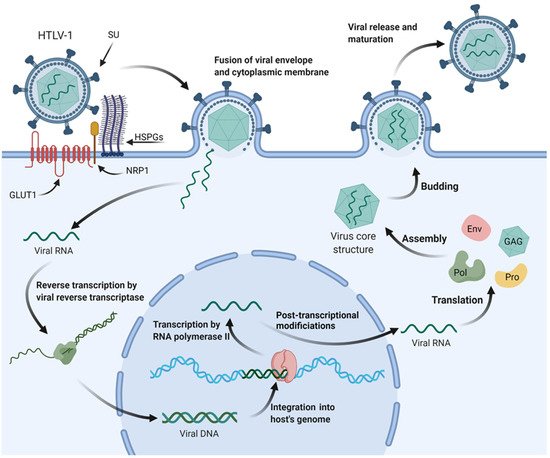
Figure 1. HTLV-1 mechanisms of cell infection and replication. Surface subunit (SU) of HTLV-1 glycoproteins interacts with heparan sulfate proteoglycans (HSPGs) of the targeted cell’s cytoplasmic membrane. A complex is then formed between the viral envelope, HSPGs, neuropilin-1 (NRP1), and glucose transporter-1 (GLUT1). The envelope fuses with the cytoplasmic membrane, and the viral RNA is released on the cytoplasm, where it will be reverse transcribed and carried to the nucleus as viral DNA for integration into the host’s genome. The provirus is then transcribed by RNA polymerase II of the cell’s transcriptional machinery, and after post-transcriptional modifications, the mature viral mRNA is transported back to the cytoplasm. Translation of viral mRNA and alternatively spliced mRNAs generate the proteins necessary for viral assembly inside the host cell, such as envelope glycoprotein (Env), polymerase (Pol), protease (Pro), and structural proteins (GAG). These proteins, alongside two copies of viral RNA, migrate to the budding site and are released from the cell’s surface to further mature into infectious viral particles following protease-dependent activity.
After infection and entry into the cell, viral RNA is reverse transcribed into viral DNA or provirus. After that, the virus can integrate into the host cell genome and exploit normal cell physiology. Viral integrase recognizes and binds to viral long terminal repeats (LTRs) in viral DNA, forming a pre-integration complex (PIC). PIC associates with the host enzyme, protein phosphatase 2A, and engages with host DNA in regions of open chromatin. In addition, HTLV-1 increases genomic instability by direct actions on the DNA, and, in turn, genomic instability can also alter the proviral genome that is often mutated or deleted, thus creating an escape from the host’s immune system [15][16][17][18][19].
During infection, there is viral transmission by cell–cell contact due to an organizing center of polarized microtubules at the cell–cell junction and a virological synapse triggered by Tax protein. From there, the HTLV-1 Gag complex, viral RNAs, and enveloped HTLV-1 virions accumulate at the synapse and migrate into the uninfected cell [20][21][22]. Another way in which viral transmission happens is by using the cell itself through proliferation. That is, the virus integrates into the host genome, and after mitosis, it is transmitted to the daughter cell [16][17].
2. Leukemogenesis Pathways to Adult T Cell Leukemia
First reported in Japan in 1977, ATL is a lymphoproliferative disease that presents as an oligoclonal or monoclonal expansion of HTLV-1-infected T cells that occurs decades after infection. ATL onset is multifactorial, involving factors related to the virus and the host’s immune and inflammatory responses [1][6].
The disease is classified according to clinical characteristics into acute, chronic, lymphoma-like, and smoldering. Its manifestations include malaise, fever, weight loss, jaundice, skin lesions, adenomegaly, hypercalcemia, elevation of lactate dehydrogenase, and clinical alterations in the number of leukocytes in peripheral blood, with the presence of atypical lymphocytes [1][23][24].
In the acute phase, it is also possible to find lymphocytes with peculiar characteristics called floral cells in the peripheral blood smear, which is a characteristic clinical finding of ATL development. The diagnosis for HTLV-1 infection is performed by ELISA and confirmed by PCR and Western blot, and a worse prognosis may be expected in the acute and lymphoma forms, being more aggressive and with a median survival of one year [17][20][25].
ATL cells’ clonal expansion is promoted by the accessory proteins of HTLV-1 and the Tax protein. Tax utilizes several mechanisms for cellular transformation, including the creation of chromosomal instability, amplification of centrosomes, abrogation of DNA repair, activation of cyclin-dependent kinases and nuclear factor-kB (NF-kB), and AKT signaling, and even silencing of TP53 checkpoints. The maintenance of the ATL transformation occurs through HBZ protein activity, in addition to this protein being essential for the establishment of persistent viral infection [20][21][26][27].
3. MicroRNAs
The molecular mechanisms of HTLV-1-mediated transformation and carcinogenesis are still unknown; however, there are studies that correlate HTLV carcinogenesis to microRNAs (miRNAs), which are endogenous 18–25 nucleotide long RNAs that play regulatory roles in cell metabolism, directing messenger RNAs (mRNA) for cleavage or translational repression [28][29]. They are involved in the maintenance of a variety of biological processes, including the cell cycle, post-transcriptional gene expression, and other processes [30][31].
MiRNAs genes can be located within the introns and exons of protein-coding genes or in intergenic regions and are transcribed by RNA Polymerase II and processed by the Drosha and Dicer enzymes [31][32][33]. Deregulation of these miRNAs is a common feature in human tumors and has been described in several types of human cancer, such as lung cancer, colorectal cancer, pancreatic endocrine, and even chronic lymphocytic leukemia [34].
Discovering miRNAs, identifying their targets, and clarifying their functions has been a critical strategy for understanding normal biological processes and their roles in disease development [35]. Furthermore, the miRNA expression pattern can be correlated with cancer, so the miRNA profile can be used as a tool for cancer diagnosis and prognosis [36]. Therefore, several studies demonstrate that the cellular expression of miRNAs is affected in HTLV-1 infected cells [37][38].
This entry is adapted from the peer-reviewed paper 10.3390/ijms23105486
References
- Delamarre, L.; Rosenberg, A.R.; Pique, C.; Pham, D.; Callebaut, I.; Dokhélar, M.C. The HTLV-I Envelope Glycoproteins: Structure and Functions. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 1996, 13, S85–S91.
- Mohanty, S.; Harhaj, E.W. Mechanisms of Oncogenesis by HTLV-1 Tax. Pathogens 2020, 9, 543.
- Martinez, M.P.; Al-Saleem, J.; Green, P.L. Comparative Virology of HTLV-1 and HTLV-2. Retrovirology 2019, 16, 21.
- Eusebio-Ponce, E.; Anguita, E.; Paulino-Ramirez, R.; Candel, F.J. HTLV-1 Infection: An Emerging Risk. Pathogenesis, Epidemiology, Diagnosis and Associated Diseases. Rev. Española Quimioter. 2019, 32, 485.
- Manns, A.; Hisada, M.; La Grenade, L. Human T-Lymphotropic Virus Type I Infection. Lancet Lond. Engl. 1999, 353, 1951–1958.
- Brites, C.; Grassi, M.F.; Quaresma, J.A.S.; Ishak, R.; Vallinoto, A.C.R. Pathogenesis of HTLV-1 Infection and Progression Biomarkers: An Overview. Braz. J. Infect. Dis. 2021, 25, 101594.
- Gessain, A.; Cassar, O. Epidemiological Aspects and World Distribution of HTLV-1 Infection. Front. Microbiol. 2012, 3, 388.
- Franchini, G.; Ambinder, R.F.; Barry, M. Viral Disease in Hematology. Hematology 2000, 2000, 409–423.
- Melamed, A.; Laydon, D.J.; Gillet, N.A.; Tanaka, Y.; Taylor, G.P.; Bangham, C.R.M. Genome-Wide Determinants of Proviral Targeting, Clonal Abundance and Expression in Natural HTLV-1 Infection. PLOS Pathog. 2013, 9, e1003271.
- Wattel, E.; Cavrois, M.; Gessain, A.; Wain-Hobson, S. Clonal Expansion of Infected Cells: A Way of Life for HTLV-I. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 1996, 13, S92–S99.
- Roucoux, D.F.; Wang, B.; Smith, D.; Nass, C.C.; Smith, J.; Hutching, S.T.; Newman, B.; Lee, T.-H.; Chafets, D.M.; Murphy, E.L. A Prospective Study of Sexual Transmission of Human T Lymphotropic Virus (HTLV)–I and HTLV-II. J. Infect. Dis. 2005, 191, 1490–1497.
- Wu, K.; Bottazzi, M.E.; De La Fuente, C.; Deng, L.; Gitlin, S.D.; Maddukuri, A.; Dadgar, S.; Li, H.; Vertes, A.; Pumfery, A.; et al. Protein Profile of Tax-Associated Complexes. J. Biol. Chem. 2004, 279, 495–508.
- Igakura, T.; Stinchcombe, J.C.; Goon, P.K.C.; Taylor, G.P.; Weber, J.N.; Griffiths, G.M.; Tanaka, Y.; Osame, M.; Bangham, C.R.M. Spread of HTLV-I between Lymphocytes by Virus-Induced Polarization of the Cytoskeleton. Science 2003, 299, 1713–1716.
- Gross, C.; Thoma-Kress, A.K. Molecular Mechanisms of HTLV-1 Cell-to-Cell Transmission. Viruses 2016, 8, 74.
- Maertens, G.N. B′-Protein Phosphatase 2A Is a Functional Binding Partner of Delta-Retroviral Integrase. Nucleic Acids Res. 2016, 44, 364.
- Tasayco-Magallanes, E.; Miranda-Ulloa, E.; Romero-Ruiz, S.; Cárdenas-Bustamante, F.; Briceño-Espinoza, R.; Yana-Calatayud, B. Evaluation of Two Brands of Test of ELISA for the Diagnosis of HTLV-1 against Peruvian Samples. Rev. Chil. Infectol. 2020, 37, 780–783.
- Cook, L.; Melamed, A.; Yaguchi, H.; Bangham, C.R. The Impact of HTLV-1 on the Cellular Genome. Curr. Opin. Virol. 2017, 26, 125–131.
- Giam, C.Z.; Semmes, O.J. HTLV-1 Infection and Adult T-Cell Leukemia/Lymphoma—A Tale of Two Proteins: Tax and HBZ. Viruses 2016, 8, 161.
- Ernzen, K.J.; Panfil, A.R. Regulation of HTLV-1 Transformation. Biosci. Rep. 2022, 42, BSR20211921.
- Pereira, W.A.; Mesquita, E.M. Vírus Linfotrópico de Células T Humana (HTLV): Doenças Associadas e Dificuldades No Diagnóstico e Tratamento. Rev. Investig. Bioméd. 2016, 8, 92–101.
- Matsuoka, M.; Jeang, K.T. Human T-Cell Leukaemia Virus Type 1 (HTLV-1) Infectivity and Cellular Transformation. Nat. Rev. Cancer 2007, 7, 270–280.
- Majorovits, E.; Nejmeddine, M.; Tanaka, Y.; Taylor, G.P.; Fuller, S.D.; Bangham, C.R.M. Human T-Lymphotropic Virus-1 Visualized at the Virological Synapse by Electron Tomography. PLoS ONE 2008, 3, e2251.
- Yoshida, N.; Weinstock, D.M. Lymphoid Neoplasia: Clinicogenetic Risk Modeling in ATL. Blood 2018, 131, 159.
- Shimoyama, M. Diagnostic Criteria and Classification of Clinical Subtypes of Adult T-Cell Leukaemia-Lymphoma. A Report from the Lymphoma Study Group (1984-87). Br. J. Haematol. 1991, 79, 428–437.
- Thorstensson, R.; Albert, J.; Andersson, S. Strategies for Diagnosis of HTLV-I and -II. Transfusion 2002, 42, 780–791.
- Brand, H.; Alves, J.G.B.; Pedrosa, F.; Lucena-Silva, N. Leucemia de Células T Do Adulto. Rev. Bras. Hematol. Hemoter. 2009, 31, 375–383.
- Franchini, G. HTLV-1 and HIV-1 “Accessory” Proteins: A Misleading Misnomer. Mol. Aspects Med. 2010, 31, 331–332.
- D’Agostino, D.M.; Zanovello, P.; Watanabe, T.; Ciminale, V. The Metwork in Normal- and HTLV-1-Transformed T Cells. Adv. Cancer Res. 2012, 113, 45–83.
- Simonson, B.; Das, S. MicroRNA Therapeutics: The Next Magic Bullet? Mini Rev. Med. Chem. 2015, 15, 467.
- Bittencourt, A.L.; Farré, L. Leucemia/Linfoma de Células T Do Adulto. An. Bras. Dermatol. 2008, 83, 351–359.
- Lu, T.X.; Rothenberg, M.E. MicroRNA. J. Allergy Clin. Immunol. 2018, 141, 1202–1207.
- Garzon, R.; Calin, G.A.; Croce, C.M. MicroRNAs in Cancer. Annu. Rev. Med. 2009, 60, 167–179.
- MacFarlane, L.-A.; Murphy, P.R. MicroRNA: Biogenesis, Function and Role in Cancer. Curr. Genom. 2010, 11, 537.
- Treiber, T.; Treiber, N.; Meister, G. Regulation of MicroRNA Biogenesis and Its Crosstalk with Other Cellular Pathways. Nat. Rev. Mol. Cell Biol. 2019, 20, 5–20.
- Di Leva, G.; Garofalo, M.; Croce, C.M. MicroRNAs in Cancer. Annu. Rev. Pathol. 2014, 9, 287–314.
- Liu, B.; Li, J.; Cairns, M.J. Identifying MiRNAs, Targets and Functions. Brief. Bioinform. 2014, 15, 1–19.
- Lee, Y.S.; Dutta, A. MicroRNAs in Cancer. Annu. Rev. Pathol. 2009, 4, 199–227.
- Moles, R.; Nicot, C. The Emerging Role of MiRNAs in HTLV-1 Infection and ATLL Pathogenesis. Viruses 2015, 7, 4047.
This entry is offline, you can click here to edit this entry!
