Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
The discovery in 2006 that loss-of-function mutations in the filaggrin gene (FLG) cause ichthyosis vulgaris and can predispose to atopic dermatitis (AD) galvanized the dermatology research community and shed new light on a skin protein that was first identified in 1981. However, although outstanding work has uncovered several key functions of filaggrin in epidermal homeostasis, a comprehensive understanding of how filaggrin deficiency contributes to AD is still incomplete, including details of the upstream factors that lead to the reduced amounts of filaggrin, regardless of genotype.
- atopic dermatitis
- filaggrin
- skin
- microbiome
1. Filaggrin: A Look in the Rear-View Mirror
Filaggrin was initially isolated from a protein fraction of the stratum corneum (SC) and identified as a basic histidine-rich protein [1]. Proteins from this fraction were shown to aggregate with keratin filaments to form macrofibrils in in vitro cell-free experiments [2]. In 1981, Dale et al. designated this class of cationic structural proteins, which associate specifically with intermediate filaments but not with other types of cytoskeletal proteins, as filaggrins (for filament aggregating proteins) [2]. They showed that filaggrins are species-distinct proteins; for example, rat and mouse filaggrins have different molecular weights (48 and 30 kDa, respectively) and different amino acid totals but nevertheless exhibit similar chemical and functional properties. The ability of filaggrin to aggregate keratin filaments into tight parallel arrays [3] has been demonstrated in many different experimental settings, with high reproducibility [4][5][6][7]. This molecular bundling confers mechanical resilience and flexibility to the SC [8]. Mouse and human filaggrin bind to each three-chain building block of the intermediate filaments, possibly through ionic interactions with the coiled-coil alpha-helical regions of the keratin filaments. The stoichiometry of the interaction has been reported as two filaggrin molecules to three intermediate (keratin) filament subunits [3][9].
Further work demonstrated that filaggrin is produced as a phosphorylated precursor, profilaggrin, of 300–500 kDa, depending on the species [10][11][12], embedded in keratohyalin granules in granular keratinocytes (KCs) [2][13][14]. Subsequently, profilaggrin undergoes a multiple-step dephosphorylation process, followed by proteolytic cleavage of short linker peptides to produce various numbers of filaggrin monomers depending on the species, e.g., 10 to 12 in humans, 16 to 20 in mice [15][16][17], and only 4 in dogs [18]. Profilaggrin is present in the stratum granulosum (SG), whereas filaggrin monomers are localized in the first layers of the SC. Of note, there is often confusion between profilaggrin and filaggrin, with some authors incorrectly assigning filaggrin to the SG. Filaggrin, together with other proteins of keratohyalin granules, forms the protein moiety of the cornified cell envelope of corneocytes [19][20][21][22][23]. In rat corneocytes, filaggrin was found to represent about 10% of the cell envelope-bound proteins [24]. Early work suggested the involvement of one or more specific phosphatases in filaggrin dephosphorylation [11][25], and in 1988, a rat 40 kDa phosphatase specific for profilaggrin was isolated [26] and later characterized as a protein phosphatase 2A-type [25]. However, data concerning specific profilaggrin phosphatases in humans are scant.
In 1993, calpain 1 was proposed as a profilaggrin protease because profilaggrin cleavage requires calcium, and calpains are calcium-dependent neutral proteases [27]. In 2009, processing of human profilaggrin by human calpain 1 was confirmed in a cell-free proteolytic assay [28]. Moreover, cathepsin L-like proteinase, but not cathepsin E, isolated from rat epidermis was shown to hydrolyze profilaggrin [29]. Work in mice showed that loss of cathepsin H, produced by an shRNA approach, leads to a dramatic reduction of filaggrin monomers, owing to defective profilaggrin processing in the epidermis [30]. The results for cathepsin D are contradictory, and the enzyme is likely not essential for profilaggrin processing in vivo [29][31][32]. Furthermore, profilaggrin endoproteinase 1 (PEP1) has been identified as a cytoplasmic enzyme that is able to digest insoluble profilaggrin purified from mouse epidermis, at least in proteolytic assays [33]. In addition, both furin and PACE4, two calcium-dependent serine proteases belonging to the same family of proprotein convertases, can cleave profilaggrin in vitro at a site between the amino terminus and the first filaggrin repeat [34]. In mice, genetic deletion of matriptase/MT-SP1, a type II transmembrane serine protease expressed in epithelial cells, including KCs, leads to failure to process profilaggrin into filaggrin monomers [35]. Moreover, it has been shown that skin-specific retroviral-like aspartic protease (SASPase) activity is indispensable for processing profilaggrin in mouse epidermis [36], a requirement which may extend to human epidermis [37][38]. SASPase cleaves the linker sequence of human profilaggrin between ‘GSFLY’ and ‘QVSTH’ [36]. Moreover, it has been proposed that lympho-epithelial Kazal type inhibitor (LEKTI), a protease inhibitor encoded by the SPINK5 gene, controls profilaggrin proteolysis. Indeed, SPINK5 knockout mice or mice with a premature stop codon in SPINK5—both are mouse models of Netherton syndrome, a severe ichthyosis due to null mutations in SPINK5—display increased amounts of mature filaggrin and reduced amounts of profilaggrin, indicating that proteolytic processing of profilaggrin is enhanced [39][40]. In human KCs, it was further shown that mesotrypsin liberates a 55-kDa N-terminal fragment of profilaggrin, hence potentially contributing to filaggrin processing as well [41]. Lastly, in transgenic mice overexpressing elastase 2, the rate of profilaggrin processing into filaggrin monomers is accelerated. In line with this, in the SG of patients with Netherton syndrome, elastase 2 is upregulated and co-localizes with profilaggrin, and profilaggrin levels are reduced [42][43][44]. Thus, many different proteases have been shown to potentially cleave profilaggrin into filaggrin monomers.
In the upper SC, filaggrin is further processed by various proteases into free amino acids. The proteolysis of filaggrin is enhanced by its prior deamination or by citrullination. The latter is a post-translational modification catalyzed by peptidylarginine deiminase enzymes, resulting in the conversion of arginine into citrulline, thereby reducing its charge (citrulline is a neutral residue, whereas arginine is positively charged) and promoting detachment of the filaggrin monomer from the aggregated keratins [45][46][47]. Histidine and glutamine are then either enzymatically or spontaneously transformed to trans-urocanic acid (UCA) and pyrrolidone carboxylic acid (PCA), respectively. These amino acids and derivatives are components of the natural moisturizing factor (NMF), together with other molecules, such as lactate, chloride and sodium ions, and urea, thus ensuring proper SC hydration [48]. Interestingly, studies with histidinemic or caspase-14-deficient mice or with epidermal equivalents knocked-down for FLG have shown a role for trans-UCA in the protection of skin against the damaging effects of UV-B radiation [5][49][50][51][52][53][54].
In 1986, Scott and Harding established that proteolysis of filaggrin in rat skin is controlled by atmospheric humidity. Filaggrin is first detected at day 20 of rat gestation in all layers of the neo-formed SC. A few hours after birth, filaggrin disappears from the upper SC layers and accumulates in the innermost half of the SC, before being confined to a thin layer at the bottom of the SC, two days after birth. In addition, keeping the newborn rat in 100% humidity or applying occlusive patches onto the skin of adult rats prevents the activation of filaggrin degradation. The authors concluded that the lowering of atmospheric humidity promotes filaggrin proteolysis into water-retaining molecules [55]. This was confirmed first in mice [56] and then in humans, using 3D epidermal equivalents [57]. Indeed, when human epidermis is produced ex vivo at low atmospheric humidity, as compared to the typical saturated conditions, profilaggrin synthesis is increased, as is filaggrin degradation into UCA and PCA. This is in line with measurements conducted in humans, showing that NMF levels are low at birth and increase within hours or days after birth in healthy baby skin [58]. These results suggest that changes in the skin microenvironment, as occurs at birth, activate filaggrin degradation but not necessarily via increased protease activity. Indeed, it has been shown in vitro that reducing the external humidity increases filaggrin deimination and, as a consequence, the molecule’s dissociation from the filamentous corneocyte matrix [46][47][57][59]. This may render filaggrin more accessible to proteases [48]. Moreover, these results suggest that improper bundling of filaggrin with keratin filaments might promote filaggrin degradation in the absence of other alterations, such as increased activity of proteases. Besides external humidity, the neutral cysteine protease bleomycin hydrolase has been shown to participate in the breakdown of deiminated filaggrin into amino acids in a cell-free proteolytic assay [28]. Moreover, bleomycin hydrolase-deficient mice display reduced amounts of skin NMF and higher levels of filaggrin [60]. Furthermore, in healthy baby skin, regional upregulation of bleomycin hydrolase activity, e.g., in the cheek when compared to the elbow flexure, might contribute to increased skin hydration at specific body sites [58].
Thus, SASPase and calpain 1 might play important roles in the processing of profilaggrin into filaggrin, whereas bleomycin hydrolase and caspase 14 are probably major proteinases responsible for filaggrin breakdown in the skin (Table 1, Figure 1); however, the contribution of other proteinases cannot be ruled out.
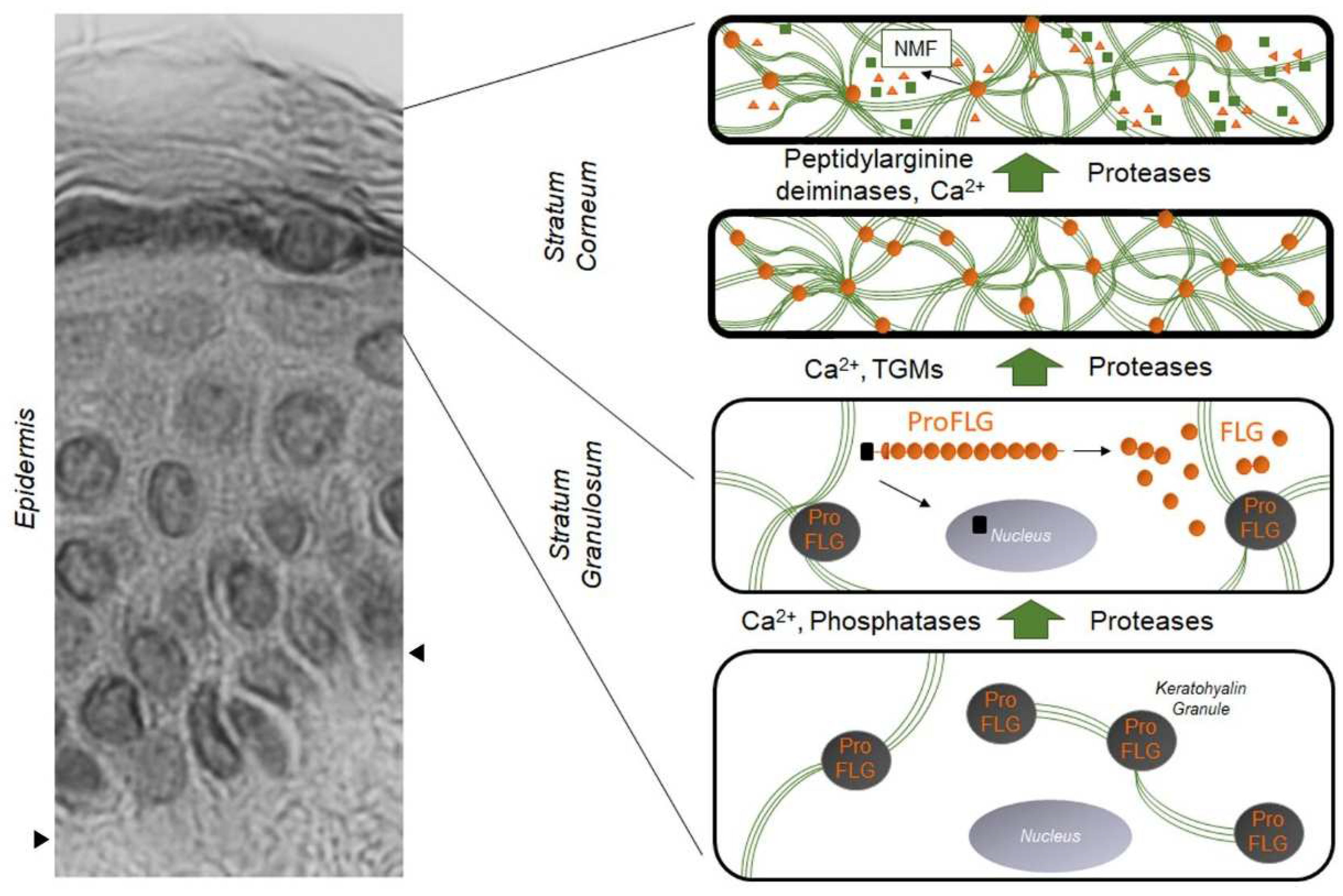
Figure 1. Evolution and distribution of profilaggrin and filaggrin during epidermal differentiation: dark gray disks containing profilaggrin are keratohyalin granules present in the cytoplasm of granular KCs. Under the action of phosphatases, proteases, and Ca2+, profilaggrin is expelled in the cytoplasm and degraded into filaggrin monomers (small orange disks). Green lines are keratin filaments aggregated by filaggrin monomers in the lower SC. Under the action of transglutaminases (TGMs), filaggrin molecules may be covalently linked to the cornified envelope. In the upper SC, filaggrin is further processed into free amino acids by the sequential action of peptidylarginine deaminases and proteases, producing the Natural Moisturizing Factor (NMF, green squares and orange triangles). The left part of the figure is a hematoxylin–eosin staining of a healthy human epidermis; arrows show the dermis–epidermis junction.
Table 1. Summary of enzymes and enzyme inhibitors involved in filaggrin metabolism (sometimes still hypothetic) in the epidermis according to data generated in vitro and in animal models versus in humans.
| In Vitro and Animal Models | in Humans | |
|---|---|---|
| Dephosphorylation of profilaggrin | Phosphatase of the protein phosphatase 2A family |
|
| Profilaggrin processing | Calpain 1, cathepsins L-like and H, PEP1, furin, PACE4, MT-SP1, mesotrypsin, SASPase, LEKTI, elastase 2 | Calpain I, SASPase, mesotrypsin |
| Filaggrin citrullination in the stratum corneum | PAD1 and/or 3 | PAD1 and/or 3 |
| Proteolysis of filaggrin in the stratum corneum | Bleomycin hydrolase, calpain 1, caspase 14 | Bleomycin hydrolase, caspase 14 |
2. The Impact of the FLG Gene: From Ichthyosis Vulgaris to Atopic Dermatitis
Ichthyosis vulgaris (IV) is a common skin disorder in humans, characterized by dry, rough, and scaly skin. In 1985, it was first reported that keratohyalin granules, profilaggrin, and filaggrin are all reduced or absent in the epidermis of patients with IV and that these biochemical abnormalities correlate with the clinical severity of the disease [61]. In 1991, high-resolution ultrastructural immunolabeling showed many small filaggrin-positive granules in association with bundles of keratin filaments in IV SG [20]. However, unequivocally, the amounts of filaggrin are much lower in the SC of IV patients compared to that of healthy volunteers [20]. In 2006, genetic studies shed new light on the IV disorder, with the identification of loss-of-function mutations in the FLG gene associated with moderate or severe IV [62]. In the same year, Palmer et al. showed that FLG loss-of-function mutations are strongly associated with atopic dermatitis (AD) [63], suggesting that low amounts of filaggrin in AD might result from genetic causes. Indeed, earlier pioneering reports showed a decrease of filaggrin in lesional AD epidermis [64], regardless of skin lesion presence or absence [65][66]. AD, also known as atopic eczema, is the most common inflammatory skin diseases, affecting 1%–36% of children and up to 18% of adults, of whom approximately 20% have moderate-to-severe disease [67][68][69][70]. Here it is important to emphasize that reduced filaggrin levels are also observed in AD patients who are genotypically the wild type for FLG [65][71], demonstrating that FLG variants are not the only factors responsible for filaggrin downregulation in AD skin, and that other factors (e.g., environmental, metabolic) are also likely to be involved. Moreover, African American patients with AD exhibit normal levels of filaggrin, in contrast to Asian and European patients [66][71]. On the other hand, pediatric AD, regardless of the patient’s ethnic background, might be characterized by normal skin levels of filaggrin [72]; however, most studies lack either FLG genotyping or measurements of filaggrin amounts [64][72][73][74].
The half-life of rodent filaggrin in the SC is 6–9 h, demonstrating that filaggrin degradation is a relatively rapid process [16][75]. This high turnover of filaggrin may confer vulnerability to the epidermis because factors that lead to reduced filaggrin expression or that accelerate its degradation could have rapid effects on the biological processes in which filaggrin is involved. Conversely, environmental factors or therapies able to augment filaggrin expression might have rapid beneficial effects. Thus, it is important to decipher accurately all the physiological roles of filaggrin in the epidermis so that pathways potentially ameliorable by filaggrin-targeted therapy are fully identified.
3. FLG Null Mutations Are Strong Genetic Factors in AD
The strong association between FLG null mutations and AD (OR = 13.4), first observed by Palmer et al. [63], was subsequently confirmed by many other research groups in studies of various ethnic populations [76][77][78][79][80][81][82][83][84][85][86][87][88][89][90][91][92][93][94][95][96][97][98][99][100][101][102]. The strength of this association was initially reported to depend on FLG genotype; that is, in a cohort of 186 patients with AD, homozygosis for the combined null genotype of six major null mutations (R501X, 2282del4, R2447X, S3247X, 3702delG, and 3673delC) was more strongly associated with the disease (OR = 85.9) compared to heterozygosis (OR = 4.6) [103]. Furthermore, Brown et al. found, in a large cohort of children, that heterozygosis is not significantly associated with AD (OR = 1.2), as opposed to homozygosis (OR = 26.9) [104]. Nomura and Kabashima suggest that the initial overestimation of the strength of the association of FLG null mutations with AD, especially in heterozygous patients, is likely due to recruitment of patients with severe disease symptoms [67]. Nevertheless, it remains that FLG homozygosis or compound heterozygosis significantly increases the risk of developing AD, at least in adult European and Asian populations [71][105]. However, Morar et al. found FLG null mutations in 26.7% of young patients with AD, but also in 14.4% of children without AD [106]. Subsequent studies have confirmed that about 40%–50% of all carriers of FLG null alleles never experience eczema [107][108]. Thus, these data show strong but incomplete penetrance of FLG variants [63][80][109][110][111], whose impact on AD development might be modulated by ethnic-specific genetic modifiers, epigenetic alterations, or other environmental factors.
Beyond FLG mutations, the number of filaggrin monomers encoded by an FLG allele can vary from 10 to 12 in humans. Individuals with 20 copies of the filaggrin monomer (i.e., homozygous 10/10) were reported to have an increased risk of developing AD, whereas those with 21 to 24 copies (genotypes 10/11 to 12/12) had no significantly elevated risk, at least in patients with moderate AD [112]. Similar work in patients with low-to-mild AD would give additional information on the power of the association between the different numbers of filaggrin monomers (i.e., genotypes) and the disease expression.
FLG null mutations have been shown to correlate with AD severity and persistence in adulthood [77][88][106][113][114]. Indeed, the observed higher prevalence of adult AD patients with an FLG null mutation (42%) [113] when compared to pediatric patients (13%) [104] argues for a role of FLG status in disease evolution, although this remains to be confirmed in large patient cohorts with different ethnic backgrounds.
A new classification of AD patients according to age, ethnicity, concomitant atopy, inflammatory milieu, SC lipid abnormalities, and FLG status designated as ‘endotypes’ has recently been proposed. In this perspective, AD patients with FLG null mutations represent a particular endotype [66][115].
This entry is adapted from the peer-reviewed paper 10.3390/ijms23105318
References
- Dale, B.A.; Lonsdale-Eccles, J.D.; Holbrook, K.A. Stratum corneum basic protein: An interfilamentous matrix protein of epidermal keratin. Curr. Probl. Dermatol. 1980, 10, 311–325.
- Dale, B.A.; Vadlamudi, B.; DeLap, L.W.; Bernstein, I.A. Similarities between stratum corneum basic protein and histidine-rich protein II from newborn rat epidermis. Biochim. Biophys. Acta 1981, 668, 98–106.
- Mack, J.W.; Steven, A.C.; Steinert, P.M. The mechanism of interaction of filaggrin with intermediate filaments. The ionic zipper hypothesis. J. Mol. Biol. 1993, 232, 50–66.
- Dale, B.A.; Presland, R.B.; Lewis, S.P.; Underwood, R.A.; Fleckman, P. Transient expression of epidermal filaggrin in cultured cells causes collapse of intermediate filament networks with alteration of cell shape and nuclear integrity. J. Investig. Dermatol. 1997, 108, 179–187.
- Pendaries, V.; Malaisse, J.; Pellerin, L.; Le Lamer, M.; Nachat, R.; Kezic, S.; Schmitt, A.M.; Paul, C.; Poumay, Y.; Serre, G.; et al. Knockdown of filaggrin in a three-dimensional reconstructed human epidermis impairs keratinocyte differentiation. J. Investig. Dermatol. 2014, 134, 2938–2946.
- Kawasaki, H.; Nagao, K.; Kubo, A.; Hata, T.; Shimizu, A.; Mizuno, H.; Yamada, T.; Amagai, M. Altered stratum corneum barrier and enhanced percutaneous immune responses in filaggrin-null mice. J. Allergy Clin. Immunol. 2012, 129, 1538–1546.
- Gruber, R.; Elias, P.M.; Crumrine, D.; Lin, T.K.; Brandner, J.M.; Hachem, J.P.; Presland, R.B.; Fleckman, P.; Janecke, A.R.; Sandilands, A.; et al. Filaggrin genotype in ichthyosis vulgaris predicts abnormalities in epidermal structure and function. Am. J. Pathol. 2011, 178, 2252–2263.
- McAleer, M.A.; Irvine, A.D. The multifunctional role of filaggrin in allergic skin disease. J. Allergy Clin. Immunol. 2013, 131, 280–291.
- Steinert, P.M.; Cantieri, J.S.; Teller, D.C.; Lonsdale-Eccles, J.D.; Dale, B.A. Characterization of a class of cationic proteins that specifically interact with intermediate filaments. Proc. Natl. Acad. Sci. USA 1981, 78, 4097–4101.
- Ramsden, M.; Loehren, D.; Balmain, A. Identification of a rapidly labelled 350K histidine-rich protein in neonatal mouse epidermis. Differentiation 1983, 23, 243–249.
- Lonsdale-Eccles, J.D.; Teller, D.C.; Dale, B.A. Characterization of a phosphorylated form of the intermediate filament-aggregating protein filaggrin. Biochemistry 1982, 21, 5940–5948.
- Meek, R.L.; Lonsdale-Eccles, J.D.; Dale, B.A. Epidermal filaggrin is synthesized on a large messenger ribonucleic acid as a high-molecular-weight precursor. Biochemistry 1983, 22, 4867–4871.
- Lonsdale-Eccles, J.D.; Haugen, J.A.; Dale, B.A. A phosphorylated keratohyalin-derived precursor of epidermal stratum corneum basic protein. J. Biol. Chem. 1980, 255, 2235–2238.
- Kuechle, M.K.; Thulin, C.D.; Presland, R.B.; Dale, B.A. Profilaggrin requires both linker and filaggrin peptide sequences to form granules: Implications for profilaggrin processing in vivo. J. Investig. Dermatol. 1999, 112, 843–852.
- Sandilands, A.; Sutherland, C.; Irvine, A.D.; McLean, W.H. Filaggrin in the frontline: Role in skin barrier function and disease. J. Cell Sci. 2009, 122, 1285–1294.
- Resing, K.A.; Walsh, K.A.; Dale, B.A. Identification of two intermediates during processing of profilaggrin to filaggrin in neonatal mouse epidermis. J. Cell Biol. 1984, 99, 1372–1378.
- Gan, S.Q.; McBride, O.W.; Idler, W.W.; Markova, N.; Steinert, P.M. Organization, structure, and polymorphisms of the human profilaggrin gene. Biochemistry 1990, 29, 9432–9440.
- Combarros, D.; Cadiergues, M.C.; Simon, M. Update on canine filaggrin: A review. Vet. Q. 2020, 40, 162–168.
- Steinert, P.M.; Marekov, L.N. The proteins elafin, filaggrin, keratin intermediate filaments, loricrin, and small proline-rich proteins 1 and 2 are isodipeptide cross-linked components of the human epidermal cornified cell envelope. J. Biol. Chem. 1995, 270, 17702–17711.
- Manabe, M.; Sanchez, M.; Sun, T.T.; Dale, B.A. Interaction of filaggrin with keratin filaments during advanced stages of normal human epidermal differentiation and in ichthyosis vulgaris. Differentiation 1991, 48, 43–50.
- Simon, M.; Sebbag, M.; Haftek, M.; Vincent, C.; Girbal-Neuhauser, E.; Rakotoarivony, J.; Sommé, G.; Schmitt, D.; Serre, G. Monoclonal antibodies to human epidermal filaggrin, some not recognizing profilaggrin. J. Investig. Dermatol. 1995, 105, 432–437.
- Simon, M.; Haftek, M.; Sebbag, M.; Montézin, M.; Girbal-Neuhauser, E.; Schmitt, D.; Serre, G. Evidence that filaggrin is a component of cornified cell envelopes in human plantar epidermis. Biochem. J. 1996, 317, 173–177.
- Albérola, G.; Schröder, J.M.; Froment, C.; Simon, M. The Amino-Terminal Part of Human FLG2 Is a Component of Cornified Envelopes. J. Investig. Dermatol. 2019, 139, 1395–1397.
- Richards, S.; Scott, I.R.; Harding, C.R.; Liddell, J.E.; Powell, G.M.; Curtis, C.G. Evidence for filaggrin as a component of the cell envelope of the newborn rat. Biochem. J. 1988, 253, 153–160.
- Kam, E.; Resing, K.A.; Lim, S.K.; Dale, B.A. Identification of rat epidermal profilaggrin phosphatase as a member of the protein phosphatase 2A family. J. Cell Sci. 1993, 106, 219–226.
- Haugen-Scofield, J.; Resing, K.A.; Dale, B.A. Characterization of an epidermal phosphatase specific for filaggrin phosphorylated by casein kinase II. J. Investig. Dermatol. 1988, 91, 553–559.
- Resing, K.A.; al-Alawi, N.; Blomquist, C.; Fleckman, P.; Dale, B.A. Independent regulation of two cytoplasmic processing stages of the intermediate filament-associated protein filaggrin and role of Ca2+ in the second stage. J. Biol. Chem. 1993, 268, 25139–25145.
- Kamata, Y.; Taniguchi, A.; Yamamoto, M.; Nomura, J.; Ishihara, K.; Takahara, H.; Hibino, T.; Takeda, A. Neutral cysteine protease bleomycin hydrolase is essential for the breakdown of deiminated filaggrin into amino acids. J. Biol. Chem. 2009, 284, 12829–12836.
- Kawada, A.; Hara, K.; Hiruma, M.; Noguchi, H.; Ishibashi, A. Rat epidermal cathepsin L-like proteinase: Purification and some hydrolytic properties toward filaggrin and synthetic substrates. J. Biochem. 1995, 118, 332–337.
- Naeem, A.S.; Tommasi, C.; Cole, C.; Brown, S.J.; Zhu, Y.; Way, B.; Willis Owen, S.A.; Moffatt, M.; Cookson, W.O.; Harper, J.I.; et al. A mechanistic target of rapamycin complex 1/2 (mTORC1)/V-Akt murine thymoma viral oncogene homolog 1 (AKT1)/cathepsin H axis controls filaggrin expression and processing in skin, a novel mechanism for skin barrier disruption in patients with atopic dermatitis. J. Allergy Clin. Immunol. 2017, 139, 1228–1241.
- Kashima, M.; Fukuyama, K.; Kikuchi, M.; Epstein, W.L. Limited proteolysis of high molecular weight histidine-rich protein of rat epidermis by epidermal proteinases. J. Investig. Dermatol. 1988, 90, 829–833.
- Egberts, F.; Heinrich, M.; Jensen, J.M.; Winoto-Morbach, S.; Pfeiffer, S.; Wickel, M.; Schunck, M.; Steude, J.; Saftig, P.; Proksch, E.; et al. Cathepsin D is involved in the regulation of transglutaminase 1 and epidermal differentiation. J. Cell Sci. 2004, 117, 2295–2307.
- Resing, K.A.; Thulin, C.; Whiting, K.; al-Alawi, N.; Mostad, S. Characterization of profilaggrin endoproteinase 1. A regulated cytoplasmic endoproteinase of epidermis. J. Biol. Chem. 1995, 270, 28193–28198.
- Pearton, D.J.; Nirunsuksiri, W.; Rehemtulla, A.; Lewis, S.P.; Presland, R.B.; Dale, B.A. Proprotein convertase expression and localization in epidermis: Evidence for multiple roles and substrates. Exp. Dermatol. 2001, 10, 193–203.
- List, K.; Szabo, R.; Wertz, P.W.; Segre, J.; Haudenschild, C.C.; Kim, S.Y.; Bugge, T.H. Loss of proteolytically processed filaggrin caused by epidermal deletion of Matriptase/MT-SP1. J. Cell Biol. 2003, 163, 901–910.
- Matsui, T.; Miyamoto, K.; Kubo, A.; Kawasaki, H.; Ebihara, T.; Hata, K.; Tanahashi, S.; Ichinose, S.; Imoto, I.; Inazawa, J.; et al. SASPase regulates stratum corneum hydration through profilaggrin-to-filaggrin processing. EMBO Mol. Med. 2011, 3, 320–333.
- Donovan, M.; Salamito, M.; Thomas-Collignon, A.; Simonetti, L.; Desbouis, S.; Rain, J.C.; Formstecher, E.; Bernard, D. Filaggrin and filaggrin 2 processing are linked together through skin aspartic acid protease activation. PLoS ONE 2020, 15, e0232679.
- Bernard, D.; Méhul, B.; Thomas-Collignon, A.; Delattre, C.; Donovan, M.; Schmidt, R. Identification and characterization of a novel retroviral-like aspartic protease specifically expressed in human epidermis. J. Investig. Dermatol. 2005, 125, 278–287.
- Descargues, P.; Deraison, C.; Bonnart, C.; Kreft, M.; Kishibe, M.; Ishida-Yamamoto, A.; Elias, P.; Barrandon, Y.; Zambruno, G.; Sonnenberg, A.; et al. Spink5-deficient mice mimic Netherton syndrome through degradation of desmoglein 1 by epidermal protease hyperactivity. Nat. Genet. 2005, 37, 56–65.
- Hewett, D.R.; Simons, A.L.; Mangan, N.E.; Jolin, H.E.; Green, S.M.; Fallon, P.G.; McKenzie, A.N. Lethal, neonatal ichthyosis with increased proteolytic processing of filaggrin in a mouse model of Netherton syndrome. Hum. Mol. Genet. 2005, 14, 335–346.
- Yamamoto-Tanaka, M.; Makino, T.; Motoyama, A.; Miyai, M.; Tsuboi, R.; Hibino, T. Multiple pathways are involved in DNA degradation during keratinocyte terminal differentiation. Cell Death. Dis. 2014, 5, e1181.
- Bonnart, C.; Deraison, C.; Lacroix, M.; Uchida, Y.; Besson, C.; Robin, A.; Briot, A.; Gonthier, M.; Lamant, L.; Dubus, P.; et al. Elastase 2 is expressed in human and mouse epidermis and impairs skin barrier function in Netherton syndrome through filaggrin and lipid misprocessing. J. Clin. Investig. 2010, 120, 871–882.
- Furio, L.; Hovnanian, A. Netherton syndrome: Defective kallikrein inhibition in the skin leads to skin inflammation and allergy. Biol. Chem. 2014, 395, 945–958.
- Barbieux, C.; Bonnet des Claustres, M.; Fahrner, M.; Petrova, E.; Tsoi, L.C.; Gouin, O.; Leturcq, F.; Nicaise-Roland, P.; Bole, C.; Béziat, V.; et al. Netherton syndrome subtypes share IL-17/IL-36 signature with distinct IFN-α and allergic responses. J. Allergy Clin. Immunol. 2022, 149, 1358–1372.
- Méchin, M.C.; Cau, L.; Galliano, M.F.; Daunes-Marion, S.; Poigny, S.; Vidaluc, J.L.; Bessou-Touya, S.; Takahara, H.; Serre, G.; Duplan, H.; et al. Acefylline activates filaggrin deimination by peptidylarginine deiminases in the upper epidermis. J. Dermatol. Sci. 2016, 81, 101–106.
- Méchin, M.C.; Takahara, H.; Simon, M. Deimination and Peptidylarginine Deiminases in Skin Physiology and Diseases. Int. J. Mol. Sci. 2020, 21, 566.
- Briot, J.; Simon, M.; Méchin, M.C. Deimination, Intermediate Filaments and Associated Proteins. Int J. Mol. Sci 2020, 21, 8746.
- Rawlings, A.V.; Harding, C.R. Moisturization and skin barrier function. Dermatol. Ther. 2004, 17 (Suppl. 1), 43–48.
- Barresi, C.; Stremnitzer, C.; Mlitz, V.; Kezic, S.; Kammeyer, A.; Ghannadan, M.; Posa-Markaryan, K.; Selden, C.; Tschachler, E.; Eckhart, L. Increased sensitivity of histidinemic mice to UVB radiation suggests a crucial role of endogenous urocanic acid in photoprotection. J. Investig. Dermatol. 2011, 131, 188–194.
- Denecker, G.; Hoste, E.; Gilbert, B.; Hochepied, T.; Ovaere, P.; Lippens, S.; Van den Broecke, C.; Van Damme, P.; D’Herde, K.; Hachem, J.P.; et al. Caspase-14 protects against epidermal UVB photodamage and water loss. Nat. Cell Biol. 2007, 9, 666–674.
- Gibbs, N.K.; Tye, J.; Norval, M. Recent advances in urocanic acid photochemistry, photobiology and photoimmunology. Photochem. Photobiol. Sci. 2008, 7, 655–667.
- Mildner, M.; Jin, J.; Eckhart, L.; Kezic, S.; Gruber, F.; Barresi, C.; Stremnitzer, C.; Buchberger, M.; Mlitz, V.; Ballaun, C.; et al. Knockdown of filaggrin impairs diffusion barrier function and increases UV sensitivity in a human skin model. J. Investig. Dermatol. 2010, 130, 2286–2294.
- Hoste, E.; Kemperman, P.; Devos, M.; Denecker, G.; Kezic, S.; Yau, N.; Gilbert, B.; Lippens, S.; De Groote, P.; Roelandt, R.; et al. Caspase-14 is required for filaggrin degradation to natural moisturizing factors in the skin. J. Investig. Dermatol. 2011, 131, 2233–2241.
- Devos, M.; Prawitt, J.; Staumont-Salle, D.; Hoste, E.; Fleury, S.; Bouchaert, E.; Gilbert, B.; Lippens, S.; Vandenabeele, P.; Dombrowicz, D.; et al. Filaggrin degradation by caspase-14 is required for UVB photoprotection but does not influence allergic sensitization in a mouse model of atopic dermatitis. J. Investig. Dermatol. 2012, 132, 2857–2860.
- Scott, I.R.; Harding, C.R. Filaggrin breakdown to water binding compounds during development of the rat stratum corneum is controlled by the water activity of the environment. Dev. Biol. 1986, 115, 84–92.
- Akiyama, K.; Senshu, T. Dynamic aspects of protein deimination in developing mouse epidermis. Exp. Dermatol. 1999, 8, 177–186.
- Cau, L.; Pendaries, V.; Lhuillier, E.; Thompson, P.R.; Serre, G.; Takahara, H.; Méchin, M.C.; Simon, M. Lowering relative humidity level increases epidermal protein deimination and drives human filaggrin breakdown. J. Dermatol. Sci. 2017, 86, 106–113.
- McAleer, M.A.; Jakasa, I.; Raj, N.; O’Donnell, C.P.F.; Lane, M.E.; Rawlings, A.V.; Voegeli, R.; McLean, W.H.I.; Kezic, S.; Irvine, A.D. Early-life regional and temporal variation in filaggrin-derived natural moisturizing factor, filaggrin-processing enzyme activity, corneocyte phenotypes and plasmin activity: Implications for atopic dermatitis. Br. J. Dermatol. 2018, 179, 431–441.
- Harding, C.R.; Scott, I.R. Histidine-rich proteins (filaggrins): Structural and functional heterogeneity during epidermal differentiation. J. Mol. Biol. 1983, 170, 651–673.
- Thyssen, J.P.; Jakasa, I.; Riethmüller, C.; Schön, M.P.; Braun, A.; Haftek, M.; Fallon, P.G.; Wróblewski, J.; Jakubowski, H.; Eckhart, L.; et al. Filaggrin Expression and Processing Deficiencies Impair Corneocyte Surface Texture and Stiffness in Mice. J. Investig. Dermatol. 2020, 140, 615–623.e5.
- Sybert, V.P.; Dale, B.A.; Holbrook, K.A. Ichthyosis vulgaris: Identification of a defect in synthesis of filaggrin correlated with an absence of keratohyaline granules. J. Investig. Dermatol. 1985, 84, 191–194.
- Smith, F.J.; Irvine, A.D.; Terron-Kwiatkowski, A.; Sandilands, A.; Campbell, L.E.; Zhao, Y.; Liao, H.; Evans, A.T.; Goudie, D.R.; Lewis-Jones, S.; et al. Loss-of-function mutations in the gene encoding filaggrin cause ichthyosis vulgaris. Nat. Genet. 2006, 38, 337–342.
- Palmer, C.N.; Irvine, A.D.; Terron-Kwiatkowski, A.; Zhao, Y.; Liao, H.; Lee, S.P.; Goudie, D.R.; Sandilands, A.; Campbell, L.E.; Smith, F.J.; et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat. Genet. 2006, 38, 441–446.
- Seguchi, T.; Cui, C.Y.; Kusuda, S.; Takahashi, M.; Aisu, K.; Tezuka, T. Decreased expression of filaggrin in atopic skin. Arch. Dermatol. Res. 1996, 288, 442–446.
- Pellerin, L.; Henry, J.; Hsu, C.Y.; Balica, S.; Jean-Decoster, C.; Méchin, M.C.; Hansmann, B.; Rodriguez, E.; Weindinger, S.; Schmitt, A.M.; et al. Defects of filaggrin-like proteins in both lesional and nonlesional atopic skin. J. Allergy Clin. Immunol. 2013, 131, 1094–1102.
- Czarnowicki, T.; He, H.; Krueger, J.G.; Guttman-Yassky, E. Atopic dermatitis endotypes and implications for targeted therapeutics. J. Allergy Clin. Immunol. 2019, 143, 1–11.
- Nettis, E.; Ortoncelli, M.; Pellacani, G.; Foti, C.; Di Leo, E.; Patruno, C.; Rongioletti, F.; Argenziano, G.; Ferrucci, S.M.; Macchia, L.; et al. A Multicenter Study on the Prevalence of Clinical Patterns and Clinical Phenotypes in Adult Atopic Dermatitis. J. Investig. Allergol. Clin. Immunol. 2020, 30, 448–450.
- Morelli, P.; Gaspari, M.; Gabriele, C.; Dastoli, S.; Bennardo, L.; Pavel, A.B.; Patruno, C.; Del Duca, E.; Nisticò, S.P. Proteomic analysis from skin swabs reveals a new set of proteins identifying skin impairment in atopic dermatitis. Exp. Dermatol. 2021, 30, 811–819.
- Deckers, I.A.; McLean, S.; Linssen, S.; Mommers, M.; van Schayck, C.P.; Sheikh, A. Investigating international time trends in the incidence and prevalence of atopic eczema 1990-2010: A systematic review of epidemiological studies. PLoS ONE 2012, 7, e39803.
- Mathiesen, S.M.; Thomsen, S.F. The prevalence of atopic dermatitis in adults: Systematic review on population studies. Dermatol. Online J. 2019, 25.
- Nomura, T.; Kabashima, K. Advances in atopic dermatitis in 2019-2020: Endotypes from skin barrier, ethnicity, properties of antigen, cytokine profiles, microbiome, and engagement of immune cells. J. Allergy Clin. Immunol. 2021, 148, 1451–1462.
- Esaki, H.; Brunner, P.M.; Renert-Yuval, Y.; Czarnowicki, T.; Huynh, T.; Tran, G.; Lyon, S.; Rodriguez, G.; Immaneni, S.; Johnson, D.B.; et al. Early-onset pediatric atopic dermatitis is T(H)2 but also T(H)17 polarized in skin. J. Allergy Clin. Immunol. 2016, 138, 1639–1651.
- Renert-Yuval, Y.; Del Duca, E.; Pavel, A.B.; Fang, M.; Lefferdink, R.; Wu, J.; Diaz, A.; Estrada, Y.D.; Canter, T.; Zhang, N.; et al. The molecular features of normal and atopic dermatitis skin in infants, children, adolescents, and adults. J. Allergy Clin. Immunol. 2021, 148, 148–163.
- He, H.; Del Duca, E.; Diaz, A.; Kim, H.J.; Gay-Mimbrera, J.; Zhang, N.; Wu, J.; Beaziz, J.; Estrada, Y.; Krueger, J.G.; et al. Mild atopic dermatitis lacks systemic inflammation and shows reduced nonlesional skin abnormalities. J. Allergy Clin. Immunol. 2021, 147, 1369–1380.
- Scott, I.R.; Harding, C.R.; Barrett, J.G. Histidine-rich protein of the keratohyalin granules. Source of the free amino acids, urocanic acid and pyrrolidone carboxylic acid in the stratum corneum. Biochim. Biophys. Acta 1982, 719, 110–117.
- Sandilands, A.; O’Regan, G.M.; Liao, H.; Zhao, Y.; Terron-Kwiatkowski, A.; Watson, R.M.; Cassidy, A.J.; Goudie, D.R.; Smith, F.J.; McLean, W.H.; et al. Prevalent and rare mutations in the gene encoding filaggrin cause ichthyosis vulgaris and predispose individuals to atopic dermatitis. J. Investig. Dermatol. 2006, 126, 1770–1775.
- Weidinger, S.; Illig, T.; Baurecht, H.; Irvine, A.D.; Rodriguez, E.; Diaz-Lacava, A.; Klopp, N.; Wagenpfeil, S.; Zhao, Y.; Liao, H.; et al. Loss-of-function variations within the filaggrin gene predispose for atopic dermatitis with allergic sensitizations. J. Allergy Clin. Immunol. 2006, 118, 214–219.
- Marenholz, I.; Nickel, R.; Rüschendorf, F.; Schulz, F.; Esparza-Gordillo, J.; Kerscher, T.; Grüber, C.; Lau, S.; Worm, M.; Keil, T.; et al. Filaggrin loss-of-function mutations predispose to phenotypes involved in the atopic march. J. Allergy Clin. Immunol. 2006, 118, 866–871.
- Ruether, A.; Stoll, M.; Schwarz, T.; Schreiber, S.; Fölster-Holst, R. Filaggrin loss-of-function variant contributes to atopic dermatitis risk in the population of Northern Germany. Br. J. Dermatol. 2006, 155, 1093–1094.
- Nomura, T.; Sandilands, A.; Akiyama, M.; Liao, H.; Evans, A.T.; Sakai, K.; Ota, M.; Sugiura, H.; Yamamoto, K.; Sato, H.; et al. Unique mutations in the filaggrin gene in Japanese patients with ichthyosis vulgaris and atopic dermatitis. J. Allergy Clin. Immunol. 2007, 119, 434–440.
- Sandilands, A.; Terron-Kwiatkowski, A.; Hull, P.R.; O’Regan, G.M.; Clayton, T.H.; Watson, R.M.; Carrick, T.; Evans, A.T.; Liao, H.; Zhao, Y.; et al. Comprehensive analysis of the gene encoding filaggrin uncovers prevalent and rare mutations in ichthyosis vulgaris and atopic eczema. Nat. Genet. 2007, 39, 650–654.
- Baurecht, H.; Irvine, A.D.; Novak, N.; Illig, T.; Bühler, B.; Ring, J.; Wagenpfeil, S.; Weidinger, S. Toward a major risk factor for atopic eczema: Meta-analysis of filaggrin polymorphism data. J. Allergy Clin. Immunol. 2007, 120, 1406–1412.
- Ekelund, E.; Liedén, A.; Link, J.; Lee, S.P.; D’Amato, M.; Palmer, C.N.; Kockum, I.; Bradley, M. Loss-of-function variants of the filaggrin gene are associated with atopic eczema and associated phenotypes in Swedish families. Acta Derm. Venereol. 2008, 88, 15–19.
- Sasaki, T.; Kudoh, J.; Ebihara, T.; Shiohama, A.; Asakawa, S.; Shimizu, A.; Takayanagi, A.; Dekio, I.; Sadahira, C.; Amagai, M.; et al. Sequence analysis of filaggrin gene by novel shotgun method in Japanese atopic dermatitis. J. Dermatol. Sci. 2008, 51, 113–120.
- Kang, T.W.; Lee, J.S.; Oh, S.W.; Kim, S.C. Filaggrin mutation c.3321delA in a Korean patient with ichthyosis vulgaris and atopic dermatitis. Dermatology 2009, 218, 186–187.
- Müller, S.; Marenholz, I.; Lee, Y.A.; Sengler, C.; Zitnik, S.E.; Griffioen, R.W.; Meglio, P.; Wahn, U.; Nickel, R. Association of Filaggrin loss-of-function-mutations with atopic dermatitis and asthma in the Early Treatment of the Atopic Child (ETAC) population. Pediatr. Allergy Immunol. 2009, 20, 358–361.
- Ching, G.K.; Hon, K.L.; Ng, P.C.; Leung, T.F. Filaggrin null mutations in childhood atopic dermatitis among the Chinese. Int J. Immunogenet. 2009, 36, 251–254.
- Brown, S.J.; Relton, C.L.; Liao, H.; Zhao, Y.; Sandilands, A.; McLean, W.H.; Cordell, H.J.; Reynolds, N.J. Filaggrin haploinsufficiency is highly penetrant and is associated with increased severity of eczema: Further delineation of the skin phenotype in a prospective epidemiological study of 792 school children. Br. J. Dermatol. 2009, 161, 884–889.
- Ma, L.; Zhang, L.; Di, Z.H.; Zhao, L.P.; Lu, Y.N.; Xu, J.; Chen, H.D.; Gao, X.H. Association analysis of filaggrin gene mutations and atopic dermatitis in Northern China. Br. J. Dermatol. 2010, 162, 225–227.
- Greisenegger, E.; Novak, N.; Maintz, L.; Bieber, T.; Zimprich, F.; Haubenberger, D.; Gleiss, A.; Stingl, G.; Kopp, T.; Zimprich, A. Analysis of four prevalent filaggrin mutations (R501X, 2282del4, R2447X and S3247X) in Austrian and German patients with atopic dermatitis. J. Eur. Acad. Dermatol. Venereol. 2010, 24, 607–610.
- Ponińska, J.; Samoliński, B.; Tomaszewska, A.; Raciborski, F.; Samel-Kowalik, P.; Walkiewicz, A.; Lipiec, A.; Piekarska, B.; Komorowski, J.; Krzych-Fałta, E.; et al. Filaggrin gene defects are independent risk factors for atopic asthma in a Polish population: A study in ECAP cohort. PLoS ONE 2011, 6, e16933.
- Chen, H.; Common, J.E.; Haines, R.L.; Balakrishnan, A.; Brown, S.J.; Goh, C.S.; Cordell, H.J.; Sandilands, A.; Campbell, L.E.; Kroboth, K.; et al. Wide spectrum of filaggrin-null mutations in atopic dermatitis highlights differences between Singaporean Chinese and European populations. Br. J. Dermatol. 2011, 165, 106–114.
- Margolis, D.J.; Apter, A.J.; Gupta, J.; Hoffstad, O.; Papadopoulos, M.; Campbell, L.E.; Sandilands, A.; McLean, W.H.; Rebbeck, T.R.; Mitra, N. The persistence of atopic dermatitis and filaggrin (FLG) mutations in a US longitudinal cohort. J. Allergy Clin. Immunol. 2012, 130, 912–917.
- Mohiuddin, M.S.; Ramamoorthy, P.; Reynolds, P.R.; Curran-Everett, D.; Leung, D.Y. Increased compound heterozygous filaggrin mutations in severe atopic dermatitis in the United States. J. Allergy Clin. Immunol. Pract. 2013, 1, 534–536.
- Margolis, D.J.; Gupta, J.; Apter, A.J.; Hoffstad, O.; Papadopoulos, M.; Rebbeck, T.R.; Wubbenhorst, B.; Mitra, N. Exome sequencing of filaggrin and related genes in African-American children with atopic dermatitis. J. Investig. Dermatol. 2014, 134, 2272–2274.
- Polcari, I.; Becker, L.; Stein, S.L.; Smith, M.S.; Paller, A.S. Filaggrin gene mutations in African Americans with both ichthyosis vulgaris and atopic dermatitis. Pediatr. Dermatol. 2014, 31, 489–492.
- Sasaki, T.; Furusyo, N.; Shiohama, A.; Takeuchi, S.; Nakahara, T.; Uchi, H.; Hirota, T.; Tamari, M.; Shimizu, N.; Ebihara, T.; et al. Filaggrin loss-of-function mutations are not a predisposing factor for atopic dermatitis in an Ishigaki Island under subtropical climate. J. Dermatol. Sci. 2014, 76, 10–15.
- Komova, E.G.; Shintyapina, A.B.; Makarova, S.I.; Ivanov, M.K.; Chekryga, E.A.; Kaznacheeva, L.F.; Vavilin, V.A. Filaggrin mutations in a Western siberian population and their association with atopic dermatitis in children. Genet. Test. Mol. Biomark. 2014, 18, 791–796.
- Park, J.; Jekarl, D.W.; Kim, Y.; Kim, J.; Kim, M.; Park, Y.M. Novel FLG null mutations in Korean patients with atopic dermatitis and comparison of the mutational spectra in Asian populations. J. Dermatol. 2015, 42, 867–873.
- Trisnowati, N.; Soebono, H.; Sadewa, A.H.; Kunisada, M.; Yogianti, F.; Nishigori, C. A novel filaggrin gene mutation 7487delC in an Indonesian (Javanese) patient with atopic dermatitis. Int. J. Dermatol. 2016, 55, 695–697.
- Margolis, D.J.; Mitra, N.; Wubbenhorst, B.; D’Andrea, K.; Kraya, A.A.; Hoffstad, O.; Shah, S.; Nathanson, K.L. Association of Filaggrin Loss-of-Function Variants With Race in Children With Atopic Dermatitis. JAMA Dermatol. 2019, 155, 1269–1276.
- González-Tarancón, R.; Sanmartín, R.; Lorente, F.; Salvador-Rupérez, E.; Hernández-Martín, A.; Rello, L.; Puzo, J.; Gilaberte, Y. Prevalence of FLG loss-of-function mutations R501X, 2282del4, and R2447X in Spanish children with atopic dermatitis. Pediatr. Dermatol. 2020, 37, 98–102.
- Brown, S.J.; Sandilands, A.; Zhao, Y.; Liao, H.; Relton, C.L.; Meggitt, S.J.; Trembath, R.C.; Barker, J.N.; Reynolds, N.J.; Cordell, H.J.; et al. Prevalent and low-frequency null mutations in the filaggrin gene are associated with early-onset and persistent atopic eczema. J. Investig. Dermatol. 2008, 128, 1591–1594.
- Brown, S.J.; Relton, C.L.; Liao, H.; Zhao, Y.; Sandilands, A.; Wilson, I.J.; Burn, J.; Reynolds, N.J.; McLean, W.H.; Cordell, H.J. Filaggrin null mutations and childhood atopic eczema: A population-based case-control study. J. Allergy Clin. Immunol. 2008, 121, 940–946.
- Gruber, R.; Janecke, A.R.; Grabher, D.; Horak, E.; Schmuth, M.; Lercher, P. Lower prevalence of common filaggrin mutations in a community sample of atopic eczema: Is disease severity important? Wien. Klin. Wochenschr. 2010, 122, 551–557.
- Morar, N.; Cookson, W.O.; Harper, J.I.; Moffatt, M.F. Filaggrin mutations in children with severe atopic dermatitis. J. Investig. Dermatol. 2007, 127, 1667–1672.
- Henderson, J.; Northstone, K.; Lee, S.P.; Liao, H.; Zhao, Y.; Pembrey, M.; Mukhopadhyay, S.; Smith, G.D.; Palmer, C.N.; McLean, W.H.; et al. The burden of disease associated with filaggrin mutations: A population-based, longitudinal birth cohort study. J. Allergy Clin. Immunol. 2008, 121, 872–877.
- Thyssen, J.P.; Carlsen, B.C.; Bisgaard, H.; Giwercman, C.; Johansen, J.D.; Linneberg, A.; Meldgaard, M.; Szecsi, P.B.; Stender, S.; Menné, T. Individuals who are homozygous for the 2282del4 and R501X filaggrin null mutations do not always develop dermatitis and complete long-term remission is possible. J. Eur. Acad. Dermatol. Venereol. 2012, 26, 386–389.
- Giardina, E.; Paolillo, N.; Sinibaldi, C.; Novelli, G. R501X and 2282del4 filaggrin mutations do not confer susceptibility to psoriasis and atopic dermatitis in Italian patients. Dermatology 2008, 216, 83–84.
- Cascella, R.; Foti Cuzzola, V.; Lepre, T.; Galli, E.; Moschese, V.; Chini, L.; Mazzanti, C.; Fortugno, P.; Novelli, G.; Giardina, E. Full sequencing of the FLG gene in Italian patients with atopic eczema: Evidence of new mutations, but lack of an association. J. Investig. Dermatol. 2011, 131, 982–984.
- Nomura, T.; Akiyama, M.; Sandilands, A.; Nemoto-Hasebe, I.; Sakai, K.; Nagasaki, A.; Ota, M.; Hata, H.; Evans, A.T.; Palmer, C.N.; et al. Specific filaggrin mutations cause ichthyosis vulgaris and are significantly associated with atopic dermatitis in Japan. J. Investig. Dermatol. 2008, 128, 1436–1441.
- Brown, S.J.; Kroboth, K.; Sandilands, A.; Campbell, L.E.; Pohler, E.; Kezic, S.; Cordell, H.J.; McLean, W.H.; Irvine, A.D. Intragenic copy number variation within filaggrin contributes to the risk of atopic dermatitis with a dose-dependent effect. J. Investig. Dermatol. 2012, 132, 98–104.
- Barker, J.N.; Palmer, C.N.; Zhao, Y.; Liao, H.; Hull, P.R.; Lee, S.P.; Allen, M.H.; Meggitt, S.J.; Reynolds, N.J.; Trembath, R.C.; et al. Null mutations in the filaggrin gene (FLG) determine major susceptibility to early-onset atopic dermatitis that persists into adulthood. J. Investig. Dermatol. 2007, 127, 564–567.
- Rice, N.E.; Patel, B.D.; Lang, I.A.; Kumari, M.; Frayling, T.M.; Murray, A.; Melzer, D. Filaggrin gene mutations are associated with asthma and eczema in later life. J. Allergy Clin. Immunol. 2008, 122, 834–836.
- Tokura, Y.; Hayano, S. Subtypes of atopic dermatitis: From phenotype to endotype. Allergol. Int. 2021.
This entry is offline, you can click here to edit this entry!
