Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Agriculture, Dairy & Animal Science
Neuropeptide Y (NPY) is one of the most abundant and ubiquitously expressed neuropeptides in both the central and peripheral nervous systems, and its regulatory effects on feed intake and appetite have been extensively studied in a wide variety of animals, including mammalian and non-mammalian species. Recent studies have shown that this neuropeptide and its receptors are expressed in various peripheral tissues; however, research investigating the distribution and function of peripherally expressed NPY in avian (non-mammalian vertebrates) species are limited.
- neuropeptide Y
- NPY receptors
- chicken
1. Structure of Neuropeptide Y
The Neuropeptide Y (NPY) system is an ancient signaling pathway, as it is found in both vertebrates and invertebrates, highlighting a potential evolutionarily conserved function [31]. Structurally similar to PYY and PP, the amino acid sequence of NPY is one of the most highly conserved neuropeptides. As shown in Table 1, there is over 90% identity in the amino acid sequence among mammalian species, and greater than 80% identity between chicken and other species (Figure 1A) [32]. Additionally, phylogenic analysis indicates that the non-mammalian species share a common ancestor that diverged from mammals in their NPY sequence [33] (Figure 1B). The molecular structure contains numerous hydrophobic interactions, as well as and N-terminal polyproline-II-like helix and a C-terminal α-helix [34]. The N-terminal portion is responsible for interactions with various receptors, as studies have shown this segment interacts with Y1 but not Y2 [35,36]. Additionally, NPY contains two translation initiation sequences, allowing for the production of both full-length and a truncated NPY, containing only peptides 17–36 [37], which can further differentially bind to receptors.
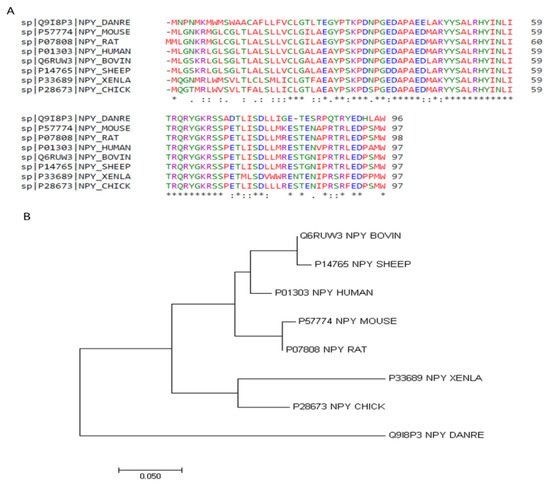
Figure 1. NPY amino acid sequence alignments (A) and phylogeny (B). Amino acid sequences were aligned using Clustal Omega 1.2.4 [32]. * positions with a single, fully conserved residue. “:” (colon) conservation between groups of strongly similar properties. “.” (period) conservation between groups of weakly similar properties. Phylogenetic tree generated with MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets [33].
Table 1. Amino acid sequence homology of NPY among several species.
| Zebrafish | Mouse | Rat | Human | Bovine | Sheep | Xenopus | Chicken | |
|---|---|---|---|---|---|---|---|---|
| Zebrafish | 100.00 | 67.71 | 67.71 | 66.67 | 65.62 | 64.58 | 62.50 | 65.62 |
| Mouse | 67.71 | 100.00 | 98.97 | 92.78 | 90.72 | 89.69 | 74.23 | 81.44 |
| Rat | 67.71 | 98.97 | 100.00 | 93.81 | 91.75 | 90.72 | 75.26 | 82.47 |
| Human | 66.67 | 92.78 | 93.81 | 100.00 | 94.85 | 93.81 | 78.35 | 84.54 |
| Bovine | 65.62 | 90.72 | 91.75 | 94.85 | 100.00 | 98.97 | 76.29 | 84.54 |
| Sheep | 64.58 | 89.69 | 90.72 | 93.81 | 98.97 | 100.00 | 75.26 | 83.51 |
| Xenopus | 62.50 | 74.23 | 75.26 | 78.35 | 76.29 | 75.26 | 100.00 | 84.54 |
| Chicken | 65.62 | 81.44 | 82.47 | 84.54 | 84.54 | 83.51 | 84.54 | 100.00 |
Numbers indicate percent identity between species, as determined by Clustal Omega 1.2.4 [26].
2. Neuropeptide Y Receptors
The physiological effects of NPY are exerted through binding to specific Y receptors, which are part of the G-protein-coupled (GPCR) family [24]. To date, 7–8 different receptors have been identified, though their presence and functionality differs among species. In mammals, Y1, Y2, Y4, Y5, and Y6 have been identified [38], whereas in fish, chicken and other avian species, Y7 is additionally present [27], and in frogs [39] and telost fish [40], Y8a and Y8b may also be present. The Y receptors have a long evolutionary history and are grouped into three subfamilies based on the homology and similarity of their amino acid sequences. The Y1 subfamily consists of Y1, Y4, Y6, and Y8, with sequence homology ranging from 40 to 60% [41]. Y1 only binds to intact NPY and PYY peptides [42,43], whereas Y4 preferentially binds PP over NPY or PYY [44]. Additionally, Y4 shows low sequence homology among different species, making it one of the most rapidly evolving GPCR known [41]. Interestingly, Y6 is the most variable in expression and functionality across species, with a complete absence in rat [45], it is truncated in many other mammals or in specific tissues [46], and present and functional in chicken [27,28,47]. The Y2 family consists of Y2 and Y7, and likely arose from a gene duplication of Y1 in an invertebrate ancestor, creating Y2 [31]. Unlike Y1, Y2 can bind truncated forms of NPY in mammals and chicken [48,49], though with less affinity in fish [50]. The Y5 subfamily consists of a single member, Y5, that similarly came from a duplication of Y1, after the creation of Y2 [31], but only has approximately 20% sequence homology with Y1 or Y2 [51]. Along with Y1, Y5 is the receptor responsible for the canonical orexigenic effects of NPY [52,53,54,55].
3. Regulation of Avian Neuropeptide Y Expression
The regulation of avian NPY expression involves nutritional, hormonal, genetic, and environmental factors. Indeed, early studies showed that negative energy conditions such as food restriction and deprivation enhance hypothalamic NPY mRNA expression [82] and neuron activity [147]. A study conducted by Zhou et al. [148] showed that chickens subjected to fasting for up to 72 h exhibited increased NPY content in the hypothalamic infundibular nucleus (IN) and paraventricular nucleus (PVN), but not in the lateral hypothalamic area (LHA). In the PVN, NPY returned to pre-fasting levels after 24 h of re-feeding. However, the level of NPY was unaffected in the IN, suggesting that fasting and re-feeding of broiler chickens can differentially affect NPY in the brain. A more recent study showed an increased NPY expression associated with lowered feed intake, particularly in 3-week-old cockerels, confirming that NPY is associated with the nutritional state of chickens [148]. Additionally, gene expression of NPY and other orexigenic molecules were up-regulated in low growth rate as compared to high growth rate cockerels, corroborating the findings reported by previous studies [149,150]. Moreover, the increase in NPY was associated with an overexpression of brain-specific homeobox protein (BSX). This confirms the requirement of BSX for physiological expression of NPY/AgRP and stimuli of hyperphagic response in avian species as demonstrated in mice [151].
In chickens divergently selected for the ratio of abdominal fatness to live weight, Dridi’s group found that the hypothalamic expression of NPY was higher in fat compared to lean bird lines under both fed and fasted conditions [152]. The same group found that the hypothalamic expression of NPY was lower in high- compared to low-feed efficient male quails, but it remained unchanged between female lines [153], indicating potential gender-dependent effects.
NPY expression is also regulated by hormonal factors such as insulin, leptin, and glucocorticoids (GCs). These peripheral hormonal signals are integrated in the hypothalamus at the arcuate nucleus of mammals or infundibular nucleus of birds [154,155]. Intracerebroventricular (ICV) injection of GCs increases feed intake in chicks [156] in a dose-dependent manner, whereas infusion of recombinant leptin over a 6 h period significantly reduced feed intake in 3-week-old broiler chickens. This effect was mediated via selective down-regulation of the hypothalamic expression of NPY [157]. Moreover, a study was conducted with the aim to evaluate the effect of dietary energy level and feeding state on the GC-induced gene expression of hypothalamic feeding-related neuropeptides, including NPY [158]. The results showed that dexamethasone (a synthetic glucocorticoid) treatment significantly increased hypothalamic NPY expression under fasting conditions. This effect was observed in chickens fed a high-fat diet but not in their counterparts receiving a low-fat diet, suggesting that the effect of peripheral GC injection on NPY expression is dependent on dietary energy concentration. The same study showed a decrease in hypothalamic NPY levels under re-feeding conditions.
ICV injection of insulin has been shown to inhibit feed intake in young chickens via the central melanocortin system [83]. Similarly, ICV injection of insulin had an anorexgenic effect on leghorn and broiler chicks [159,160], indicating that insulin in birds, like mammals, is an anorexigenic neuropeptide. More recent studies have further explored the interaction between NPY and insulin, and have indicated that the hypophagic effect of insulin is likely mediated by the Y1 and Y2 receptors [161,162].
Environmental conditions may also alter NPY expression; however, the data are controversial and a matter of debate. For instance, heat-stressed chickens showed increased NPY mRNA [163,164], decreased NPY mRNA [125], or no change when compared to thermoneutral controls [165], though these effects may differ based on age and strain of the birds studied, as well as the temperature and duration of the heat stress. Similarly, ICV injection of NPY during heat exposure diminished the orexigenic response of broiler chicks to NPY [131], while in layer-type chicks, the response to ICV NPY was similar between thermoneutral and heat-stressed chicks [129]. Moreover, NPY treatment has been shown to exert a hypothermic effect on layer-type chickens [129,166], however this has yet to be explored in broilers, but does suggest that NPY additionally inhibits energy expenditure. The reduction in NPY abundance and function observed in some studies is a plausible explanation for the decrease in food intake during heat stress, whereas the increase reported by other authors may be induced by the increase in plasma corticosterone under stressful conditions.
This entry is adapted from the peer-reviewed paper 10.3390/vetsci9040171
This entry is offline, you can click here to edit this entry!
