The DNA damage response and repair (DDR/R) network, a sum of hierarchically structured signaling pathways that recognize and repair DNA damage, and the immune response to endogenous and/or exogenous threats, act synergistically to enhance cellular defence. On the other hand, a deregulated interplay between these systems underlines inflammatory diseases including malignancies and chronic systemic autoimmune diseases, such as systemic lupus erythematosus, systemic sclerosis, and rheumatoid arthritis. Patients with these diseases are characterized by aberrant immune response to self-antigens with widespread production of autoantibodies and multiple-tissue injury, as well as by the presence of increased oxidative stress. Recent data demonstrate accumulation of endogenous DNA damage in peripheral blood mononuclear cells from these patients, which is related to (a) augmented DNA damage formation, at least partly due to the induction of oxidative stress, and (b) epigenetically regulated functional abnormalities of fundamental DNA repair mechanisms. Because endogenous DNA damage accumulation has serious consequences for cellular health, including genomic instability and enhancement of an aberrant immune response, these results can be exploited for understanding pathogenesis and progression of systemic autoimmune diseases, as well as for the development of new treatments.
- DNA damage response and repair network
- immune response
- autoimmunity
- systemic lupus erythematosus
- systemic sclerosis
- rheumatoid arthritis
- oxidative stress
- abasic sites
- chromatin organization
- apoptosis
The DNA damage response and repair (DDR/R) network
The DNA damage response and repair (DDR/R) network is a hierarchically structured mechanism, consisting of sensors, mediators, transducers, and effectors, which recognize any defects during the cell cycle and assign the proper repair process [1]. In case of unrepaired lesions and depending on the extent and type of damage, the cell either passes the mutated genome to its offspring or is neutralized by programmed cell death (apoptosis) or senescence [2]. To compensate for the many types of DNA damage that occur, cells have developed six major DNA repair mechanisms wherein each corrects a different subset of lesions.
Nucleotide Excision Repair (NER)
NER is a fundamental DNA repair mechanism involved in the removal of bulky, helix-distorting lesions from DNA[3]. DNA adducts that are repaired by NER include cyclobutane pyrimidine dimers (CPDs) and 6-4 photoproducts (6-4 PPs) produced by UV radiation, DNA lesions generated by ROS or endogenous lipid peroxidation products, intrastrand cross-links and adducts produced by genotoxic drugs (melphalan, cisplatin), or environmental carcinogens (benzo[a]pyrene)[4][5]. There are two subpathways of NER, termed GGR (global genome repair) and TCR (transcription-coupled repair), where approximately 30 proteins are involved in both subpathways. The biological importance of NER for human health is obvious by the fact that defects in this repair pathway cause several human genetic disorders, including Cockayne syndrome (CS), xeroderma pigmentosum (XP), and trichothiodystrophy (TTD), which are all associated with photosensitivity[6].
Base Excision Repair (BER)
BER is a conserved and ubiquitous DNA repair pathway, which recognizes and removes damaged DNA bases that do not significantly distort the structure of the DNA helix[7]. BER is used by the cell to correct DNA lesions that occur through the spontaneous deamination or hydroxylation of bases and by oxidation of nucleotides by ROS produced either by normal metabolism or environmental stresses such as smoking, oxidizing chemicals, or ionizing radiation[8]. In addition, BER is implicated in the repair of alkylated DNA bases generated by endogenous or exogenous factors (carcinogens, antineoplastic drugs, etc.), which if left unrepaired produce mutations in the cells[9]. BER consists of two subpathways, known as single-nucleotide or short-patch and long-patch.
Mismatch Repair (MMR)
MMR mechanism removes base substitution and insertion/deletion mismatches that arise as a result of replication errors escaping the proofreading function of DNA polymerases[10]. Deficiencies in MMR lead to microsatellite instability (MSI), which is a pattern of hypermutation that occurs at genomic microsatellites, and is associated with unique clinical features, such as prognosis and response to therapy, and immune checkpoint blockade[11][12].
Double-Strand Breaks (DSBs) Repair
DSBs may occur as a result of exposure to both exogenous factors, including ionizing radiation, UV light and genotoxic drugs[13] and endogenous events, including oxidative stress, replication fork collapse, and telomere erosion[14]. These lesions, also, occur as programmed events during meiosis, as well as during V(D)J recombination and class-switch recombination (CSR)[15]. To maintain genomic integrity, cells have evolved several pathways to remove DSBs.
Homologous Recombination Repair (HRR)
HRR is an error-free DNA repair mechanism, which operates during the S and G2 phases of the cell cycle so that it can find a large area of homology on a sister chromatid to use as a template for resynthesizing damaged or lost bases[16].
Canonical Non-Homologous End Joining (c-NHEJ)
c-NHEJ is an error-prone process, which is active throughout the entire cell cycle. C-NHEJ is initiated by the binding of the heterodimeric protein complex X-ray repair cross complementing 5/6 to both DNA ends. Then, DNA-dependent protein kinase (DNA-PK) is recruited, a DNA dependant protein kinase, which activates X-ray repair cross-complementing protein 4 (XRCC4)-ligase IV complex to link the broken DNA ends together. However, before re-ligation, MRN complex (MRE11-Rad50-NBS1), together with the Flap Endonuclease 1 (FEN1) and Artemis, are involved in processing DNA ends[17] [18].
Alternative Non-Homologous End Joining (Alt-NHEJ)
Alt-NHEJ is a mechanistically distinct pathway of DSB repair that is frequently termed microhomology-mediated end-joining[19]. Indeed, the foremost distinguishing property of alt-NHEJ is the use of 5–25 base pair microhomologous sequences during the alignment of broken ends before joining, thereby resulting in deletions flanking the original break[20].
Single Strand Annealing (SSA)
This process involves a DSB between homologous repeats, followed by DSB end resection that generates 3′-ssDNA, which reveals flanking homologous sequences that are annealed together to form a synapsed intermediate. This intermediate is then processed for ligation, which requires endonucleolytic cleavage of nonhomologous 3′-ssDNA tails, and polymerase filling of the gaps[21].
Interstrand Cross-Link (ICL) Repair
The formation of cross-links between the two strands of DNA is considered a critical event, causing cell cycle and replication arrest and eventually cell death if not repaired[22]. Cross-linking agents are exogenous chemicals, including the drugs cyclophosphamide, melphalan, cisplatin, mitomycin C, and psoralen[23], as well as endogenously formed aldehydes [24]. There are three routes for cross-link detection in mammalian cells. Adducts can be recognized in otherwise unperturbed duplex DNA by factors that recognize DNA damage, via encounter with the transcription machinery or by blocking a replication fork, triggering a repair response that would remove the cross-link and restore replication[25].
Direct Repair Pathway
The direct repair mechanism is a single step pathway[26]. The sole protein involved, O6-methylguanine-DNA methyltransferase (MGMT), removes alkyl groups of thymine, such as those generated by treatment with alkylating drugs, and transfers it to an internal cysteine residue of MGMT [27].
The Interplay between the DDR/R Network and the Immune Response: The Role of Oxidative Stress
The interplay between DDR/R and innate immune response has been suggested by a few studies in the recent years. Recent studies have demonstrated that deregulation of DNA repair mechanisms result in the accumulation of cytosolic single-stranded DNAs and double-stranded DNAs that can act as potent immunostimulators through the induction of the cGAS-STING-IRF3 pathway and the production of type I interferon.[28][29][30][31][32][33][34][35][36] On the contrary, loss of immune homeostasis and prolonged inflammatory response can lead to DNA damage and activate the DDR/R network, thus indicating a bi-directional relationship between DDR/R and immune response[1][37][38][39][40][41][42][43].
Systemic autoimmune diseases are characterized by aberrant immune response to self-antigens with widespread production of autoantibodies and multiple tissue injury, as well as by oxidative stress along with the excess production of reactive oxygen species (ROS) and reactive nitrogen species (RNS). The inappropriate activation of adaptive immunity and production of autoantibodies has been classically linked to autoimmunity, whereas innate immune activation and, specifically, the recognition of nucleic acids by Toll-like receptors and other cytoplasmic innate immune receptors, are gaining attention as part of the pathophysiology of autoimmune diseases[44].
DNA Double-Strand Breaks Per Se Induce Innate Immune Activation
The presence of DSBs per se has been shown to induce type I IFN production. Treatment of healthy donor-derived primary monocytes with etoposide, mitomycin C or adriamycin, three DSB-inducing drugs, was able to induce type I and III IFNs in primary monocytes and various cell lines, suggesting that DDR-induced IFN expression is a universal mechanism that may underline different pathological processes[30]. Furthermore, basic components of DSB repair were shown to be responsible for the production of cytoplasmic ssDNA, which seems to be the main immunostimulant[35].
Defective DNA Repair and Chronic Low Level DNA Damage “Prime” the Innate Immune Response
Important data supporting that defective DNA repair primes innate immune response comes from ataxia-telangiectasia (AT), a neurodegenerative disorder associated with mutations of the central DNA repair kinase ATM[31]. In summary, ATM deficiency leads to the accumulation of DNA damage, exportation of damaged ssDNA and dsDNA into the cytoplasm, activation of the CGAS-STING pathway, and finally type I IFN production that primed cells for response to exogenous or endogenous stimuli such as viral or bacterial infections. Moreover, Günther and colleagues suggested that defective ribonucleotide removal and accumulation of base lesions and low-grade DNA damage “primed” ImmR[45].
Oxidative Stress Causes DNA Damage That Activates the Immune System
Cellular oxidative damage is a general mechanism of cell and tissue injury, which is primarily caused by free radicals and ROS[46]. ROS are produced by both endogenous and exogenous sources. Endogenous sources include the generation of ROS from mitochondria; peroxisomes, activated inflammatory cells and during the metabolism of xenobiotics. As for the extracellular sources of ROS, these include the ultraviolet A (UVA) light, chemotherapeutics, environmental toxins, and other pollutants[47][46]. DNA lesions associated with ROS are oxidized purines and pyrimidines, SSBs, DSBs, and abasic sites[48]. On exposure to oxidative stress, cells initiate a variety of defence mechanisms, including both enzymatic (superoxide dismutase, glutathione peroxidase, glutathione reductase, glutathione-S-transferase, and catalase) and non-enzymatic antioxidants [ascorbic acid (vitamin C), α-tocopherol (vitamin E), total thiol, glutathione, carotenoids, and flavonoids][49].
Οxidative stress can participate in the pathogenesis, progression, and complication of many diseases, including cancer and systemic autoimmune diseases[46]. Especially with regard to systemic autoimmune diseases, several studies have shown that SLE patients are characterized by increased oxidative stress, resulting in immune system dysregulation, abnormal activation and processing of cell-death signals, and autoantibody production[50]. In addition, oxidative stress is involved in the pathogenesis of SSc[51]. That is, SSc patients are characterized by increased production of ROS in the skin, visceral fibroblasts, and endothelial cells, as well as by reduced concentrations of various antioxidants, including antioxidant vitamins and minerals[52].Oxidative stress signs, such as intracellular ROS, lipid peroxidation, protein oxidation, DNA damage, and deregulated antioxidant defence system of the body, have also been observed in patients with RA[53].
The DDR/R Network in Systemic Autoimmune Diseases
Systemic Lupus Erythematosus
The first hint that abnormalities in DDR/R pathway may be involved in SLE pathophysiology comes from the increased frequency of polymorphisms of central molecules involved in the DDR/R pathway such as TREX1[54]. Moreover, autoantibodies against components of the DDR pathway have been detected in approximately 10%–20% of patients with SLE [55]. Some known targets of autoantibodies in SLE are the two subunits of Ku protein (Ku70 and Ku80), DNA ligase IV, XRCC4, DNA-PK, PARP, Mre11 and Werner protein[56][57].
Peripheral blood mononuclear cells (PBMCs) from SLE patients display defects in two main DNA repair pathways, namely, NER and DSB repair. SLE patients with nephritis have approximately 3–5 times higher intrinsic DNA damage compared with healthy controls. Of interest, patients with quiescent disease also exhibited increased levels of DNA damage, although lower than patients with nephritis[28][29].
Recent studies suggest that either dysregulated apoptosis or defects in dead cell clearance contribute to the perpetuation of autoimmunity and SLE pathogenesis[58]. Studies revealed a significantly higher percentage of apoptotic cells in SLE patients than in controls, which was also positively correlated with the number of plasmacytoid dendritic cells, the major type I IFN-a producer[59]. Interestingly, our previous studies have shown that genotoxic drug-induced apoptosis rates were higher in PBMCs from quiescent SLE patients than healthy controls and correlated inversely with DNA repair efficiency, supporting the hypothesis that accumulation of DNA damage contributes to increased apoptosis[28][29].
Systemic Sclerosis
Oxidative stress has been implicated in the development and perpetuation of SSc[60]. In fibroblasts isolated from the skin of patients with diffuse SSc, levels of ROS and type I collagen are significantly higher and the amounts of free thiol are significantly lower when compared to normal fibroblasts[61]. Moreover, sera from patients with diffuse SSc and lung fibrosis contain elevated levels of advanced oxidation protein products (AOPPs) compared to sera from healthy individuals or from patients with limited SSc and no lung fibrosis[62]. Furthermore, increased DNA damage levels have also been detected in the peripheral blood of patients with SSc, regardless of disease subtype (diffuse or limited SSc) or treatment[63].
Rheumatoid Arthritis
The role of oxidative DNA damage and aberrations of the DDR/R network have been long studied in RA[64]. P53 mutations and overexpression were characteristically detected in the synovium of patients with RA[65][66][67]. Immunohistochemical analysis of RA synovial tissues revealed compensatory up-regulation of MMR enzymes, especially in the synovial lining, which, however, did not completely invert the observed oxidative damage[68]. Moreover, increased endogenous DNA damage levels in peripheral blood (PBMCs or granulocytes) are observed in patients with RA compared to healthy controls[69][70][53].
Conclusion
In conclusion, the DDR/R network and the ImmR act synergistically to ensure genomic stability and cell homeostasis, therefore a balance shift in DDR/R may negatively affect ImmR and the opposite may also occur.
As depicted in Figure 1, we propose that epigenetically regulated functional abnormalities of DNA repair mechanisms (i.e., downregulation of DDR/R-related genes and condensed chromatin structure that result in defective repair) and increased endogenous DNA damage formation, partly due to the induction of oxidative stress, may result in the augmented accumulation of DNA damage (both SSBs and DSBs) in patients with systemic autoimmune diseases. This accumulation may trigger the induction of apoptosis, which facilitates autoantibody production, as well as the generation of damaged cytosolic DNA and micronuclei that both can act as potent immunostimulators through the induction of the type I IFN pathway, leading to systemic autoimmune disease expression. Notably, some of the components cGAS-STING-IRF3 pathway and the production of type I IFN, leading to systemic autoimmune disease are partially reversible following histone hyperacetylation by HDAC inhibitors.
Recent data have shown that treatment of human SLE-derived PBMCs with the HDACi vorinostat results in hyperacetylation of histone H4, chromatin decondensation, restoration of the DNA repair capacity, and decreased apoptosis rates[29]. These results are in line with previous data, showing that HDACi ameliorate disease in lupus mouse models[71][72][73], as well treatment of lupus-prone Mrl/lpr mice with the HDACi panobinostat significantly reduced circulating naïve B and plasma cell numbers and the levels of autoantibodies[74]. More importantly, in children with systemic-onset juvenile idiopathic arthritis, the HDACi givinostat was found to be safe and beneficial, particularly in reducing the arthritic features, suggesting that HDACi may have important clinical applications in the treatment of systemic autoimmunity[75].
Taken together, the results reviewed herein suggest that the deregulated interplay between DDR/R and ImmR plays a crucial role in the pathogenesis and progression of systemic autoimmune diseases. Thus, unraveling the molecular mechanisms of this interplay can be exploited for understanding pathogenesis and progression of these diseases, as well as to discover new treatment opportunities in the field.
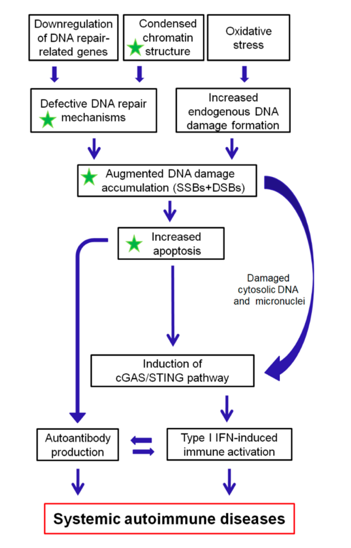
Figure 1: A proposed model of systemic autoimmune disease promotion by epigenetically regulated functional abnormalities of the DNA damage response and repair (DDR/R) network and oxidative stress. The green asterisk denotes partial reversibility following histone hyperacetylation. SSBs: single-strand breaks, DSBs: double-strand breaks.
This entry is adapted from the peer-reviewed paper 10.3390/ijms21010055
References
- Ioannis S. Pateras; Sophia Havaki; Xenia Nikitopoulou; Konstantinos Vougas; Paul A. Townsend; Michalis I. Panayiotidis; Alexandros G. Georgakilas; Vassilis G. Gorgoulis; The DNA damage response and immune signaling alliance: Is it good or bad? Nature decides when and where. Pharmacology & Therapeutics 2015, 154, 36-56, 10.1016/j.pharmthera.2015.06.011.
- Stephen Philip Jackson; Jiri Bartek; The DNA-damage response in human biology and disease.. Nature 2009, 461, 1071-8, 10.1038/nature08467.
- Sarah C. Shuck; Emily A. Short; John J. Turchi; Eukaryotic nucleotide excision repair: from understanding mechanisms to influencing biology.. Cell Research 2008, 18, 64-72, 10.1038/cr.2008.2.
- Palak Shah; Yu-Ying He; Molecular regulation of UV-induced DNA repair.. Photochemistry and Photobiology 2015, 91, 254-64, 10.1111/php.12406.
- Schärer, O.D.; Nucleotide excision repair in eukaryotes. Cold Spring Harb. Perspect. Biol. 2013, 5, a012609, .
- James E. Cleaver; Ernest T. Lam; Ingrid Revet; Disorders of nucleotide excision repair: the genetic and molecular basis of heterogeneity. Nature Reviews Genetics 2009, 10, 756-768, 10.1038/nrg2663.
- Zharkov, D.O.; Base excision DNA repair. Cell. Mol. Life Sci. 2008, 65, 1544–1565, .
- Krokan, H.E.; Bjørås, M.; Base excision repair. . Cold Spring Harb. Perspect. Biol. 2013, 5, a012583, .
- Meihua Luo; Hongzhen He; Mark R. Kelley; Millie M. Georgiadis; Redox Regulation of DNA Repair: Implications for Human Health and Cancer Therapeutic Development. Antioxidants & Redox Signaling 2010, 12, 1247-1269, 10.1089/ars.2009.2698.
- Sarah A. Martin; Christopher J. Lord; Alan Ashworth; Therapeutic Targeting of the DNA Mismatch Repair Pathway. Clinical Cancer Research 2010, 16, 5107-5113, 10.1158/1078-0432.ccr-10-0821.
- Ivan Diaz-Padilla; Nuria Romero; Eitan Amir; Xavier Matias-Guiu; Eduardo Vilar; Franco Muggia; Jesus Garcia-Donas; Mismatch repair status and clinical outcome in endometrial cancer: A systematic review and meta-analysis. Critical Reviews in Oncology/Hematology 2013, 88, 154-167, 10.1016/j.critrevonc.2013.03.002.
- Ying-Hua Li; Xiaoxu Wang; Yunfeng Pan; Ng-Hyun Lee; Dipanjan Chowdhury; Alec C. Kimmelman; Inhibition of Non-Homologous End Joining Repair Impairs Pancreatic Cancer Growth and Enhances Radiation Response. PLOS ONE 2012, 7, e39588, 10.1371/journal.pone.0039588.
- Alberto Ciccia; Stephen J. Elledge; The DNA Damage Response: Making it safe to play with knives. Molecular Cell 2010, 40, 179-204, 10.1016/j.molcel.2010.09.019.
- J. Ren; X. Liao; M. D. Vieson; M. Chen; R. Scott; J. Kazmierczak; X. M. Luo; C. M. Reilly; Inhibition of non-homologous end joining repair impairs pancreatic cancer growth and enhances radiation response. Clinical & Experimental Immunology 2017, 191, 19-31, 10.1111/cei.13046.
- Shaun P. Scott; Tej K. Pandita; The cellular control of DNA double-strand breaks.. Journal of Cellular Biochemistry 2006, 99, 1463-75, 10.1002/jcb.21067.
- Thomas Helleday; Homologous recombination in cancer development, treatment and development of drug resistance. Carcinogenesis 2010, 31, 955-960, 10.1093/carcin/bgq064.
- Anirban Chakraborty; Nisha Tapryal; Tatiana Venkova; Nobuo Horikoshi; Raj K. Pandita; Altaf H. Sarker; Partha S. Sarkar; Tej K. Pandita; Tapas K. Hazra; Classical non-homologous end-joining pathway utilizes nascent RNA for error-free double-strand break repair of transcribed genes. Nature Communications 2016, 7, 13049, 10.1038/ncomms13049.
- Anthony J. Davis; David J. Chen; DNA double strand break repair via non-homologous end-joining. Translational Cancer Research 2013, 2, 130-143, 10.3978/j.issn.2218-676X.2013.04.02.
- Iliakis, G.; Murmann, T.; Soni, A. Alternative end-joining repair pathways are the ultimate backup for abrogated classical non-homologous end-joining and homologous recombination repair: Implications for the formation of chromosome translocations. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2015, 793, 166–175.
- Amy Marie Yu; Mitch McVey; Synthesis-dependent microhomology-mediated end joining accounts for multiple types of repair junctions.. Nucleic Acids Research 2010, 38, 5706-17, 10.1093/nar/gkq379.
- Bhargava, R.; Onyango, D.O.; Stark, J.M. Regulation of Single-Strand Annealing and its Role in Genome Maintenance. Trends Genet. 2016, 32, 566–575
- Satoru Hashimoto; Hirofumi Anai; Katsuhiro Hanada; Mechanisms of interstrand DNA crosslink repair and human disorders.. Genes and Environment 2016, 38, 9, 10.1186/s41021-016-0037-9.
- Andrew J. Deans; Stephen C. West; DNA interstrand crosslink repair and cancer.. Nature Cancer 2011, 11, 467-80, 10.1038/nrc3088.
- Juan I. Garaycoechea; Gerry P. Crossan; Frederic Langevin; Maria Daly; Mark J. Arends; Ketan J. Patel; Genotoxic consequences of endogenous aldehydes on mouse haematopoietic stem cell function. Nature 2012, 489, 571-575, 10.1038/nature11368.
- Yucai Wang; Justin W. Leung; Yingjun Jiang; Megan G. Lowery; Do Huong; Karen M. Vasquez; Junjie Chen; Weidong Wang; Lei Li; FANCM and FAAP24 Maintain Genome Stability via Cooperative as well as Unique Functions. Molecular Cell 2013, 49, 997-1009, 10.1016/j.molcel.2012.12.010.
- Bernd Kaina; Markus Christmann; Steffen Naumann; Wynand P. Roos; MGMT: Key node in the battle against genotoxicity, carcinogenicity and apoptosis induced by alkylating agents. DNA Repair 2007, 6, 1079-1099, 10.1016/j.dnarep.2007.03.008.
- Hiddinga, B.I.; Pauwels, P.; Janssens, A.; van Meerbeeck, J.P. O6-Methylguanine-DNA methyltransferase (MGMT): A drugable target in lung cancer? Lung Cancer (Amst. Neth.) 2017, 107, 91–99.
- V L Souliotis; P P Sfikakis; Increased DNA double-strand breaks and enhanced apoptosis in patients with lupus nephritis. Lupus 2014, 24, 804-815, 10.1177/0961203314565413.
- Vassilis L. Souliotis; Konstantinos Vougas; Vassilis G. Gorgoulis; Petros P. Sfikakis; Defective DNA repair and chromatin organization in patients with quiescent systemic lupus erythematosus.. Arthritis Research & Therapy 2016, 18, 182, 10.1186/s13075-016-1081-3.
- Brzostek-Racine, S.; Gordon, C.; Van Scoy, S.; Reich, N.C. The DNA damage response induces IFN. J. Immunol. 2011, 187, 5336–5345
- Anetta Hartlova; Saskia F. Erttmann; Faizal Am. Raffi; Anja M. Schmalz; Ulrike Resch; Sharath Anugula; Stefan Lienenklaus; Lisa M. Nilsson; Andrea Kröger; Jonas A. Nilsson; et al. DNA Damage Primes the Type I Interferon System via the Cytosolic DNA Sensor STING to Promote Anti-Microbial Innate Immunity. Immunity 2015, 42, 332-343, 10.1016/j.immuni.2015.01.012.
- Lorenzo Galluzzi; Aitziber Buqué; Oliver Kepp; Laurence Zitvogel; Guido Kroemer; Immunological Effects of Conventional Chemotherapy and Targeted Anticancer Agents. Cancer Cell 2015, 28, 690-714, 10.1016/j.ccell.2015.10.012.
- Shen, Y.J.; Le Bert, N.; Chitre, A.A.; Koo, C.X.; Nga, X.H.; Ho, S.S.W.; Khatoo, M.; Tan, N.Y.; Ishii, K.J.; Gasser, S. Genome-derived cytosolic DNA mediates type I interferon-dependent rejection of B cell lymphoma cells. Cell Rep. 2015, 11, 460–473.
- Rania Nakad; Björn Schumacher; DNA Damage Response and Immune Defense: Links and Mechanisms. Frontiers in Genetics 2016, 7, 1065, 10.3389/fgene.2016.00147.
- Erkin Erdal; Syed Haider; Jan Rehwinkel; Adrian L. Harris; Peter J. McHugh; A prosurvival DNA damage-induced cytoplasmic interferon response is mediated by end resection factors and is limited by Trex1. Genes & Development 2017, 31, 353-369, 10.1101/gad.289769.116.
- Mackenzie, K.J.; Carroll, P.; Lettice, L.; Tarnauskaite ̇, Ž.; Reddy, K.; Dix, F.; Revuelta, A.; Abbondati, E.; Rigby, R.E.; Rabe, B.; et al. Ribonuclease H2 mutations induce a cGAS/STING-dependent innate immune response. EMBO J. 2016, 35, 831–844
- Meira, L.B.; Bugni, J.M.; Green, S.L.; Lee, C.-W.; Pang, B.; Borenshtein, D.; Rickman, B.H.; Rogers, A.B.; Moroski-Erkul, C.A.; McFaline, J.L. DNA damage induced by chronic inflammation contributes to colon carcinogenesis in mice. J. Clin. Invest. 2008, 118, 2516–2525
- Shiho Ohnishi; Ning Ma; Raynoo Thanan; Somchai Pinlaor; Olfat Hammam; Mariko Murata; Shosuke Kawanishi; DNA Damage in Inflammation-Related Carcinogenesis and Cancer Stem Cells. Oxidative Medicine and Cellular Longevity 2013, 2013, 1-9, 10.1155/2013/387014.
- Pálmai-Pallag, T.; Bachrati, C.Z. Inflammation-induced DNA damage and damage-induced inflammation: A vicious cycle. Microbes Infect. 2014, 16, 822–832
- Kidane, D.; Chae, W.J.; Czochor, J.; Eckert, K.A.; Glazer, P.M.; Bothwell, A.L.; Sweasy, J.B. Interplay between DNA repair and inflammation, and the link to cancer. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 116–139.
- Fabrícia Lima Fontes; Daniele Maria Lopes Pinheiro; Ana Helena Sales De Oliveira; Rayssa Karla De Medeiros Oliveira; Tirzah Braz Petta Lajus; Lucymara Fassarella Agnez-Lima; Role of DNA repair in host immune response and inflammation. Mutation Research/Reviews in Mutation Research 2015, 763, 246-257, 10.1016/j.mrrev.2014.11.004.
- Orsolya Király; Guanyu Gong; Werner Olipitz; Sureshkumar Muthupalani; Bevin P. Engelward; Inflammation-Induced Cell Proliferation Potentiates DNA Damage-Induced Mutations In Vivo. PLOS Genetics 2015, 11, e1004901, 10.1371/journal.pgen.1004901.
- Ana Neves-Costa; Luis F. Moita; Modulation of inflammation and disease tolerance by DNA damage response pathways. The FEBS Journal 2017, 284, 680-698, 10.1111/febs.13910.
- Argyrios N. Theofilopoulos; Dwight H. Kono; Roberto Baccala; The multiple pathways to autoimmunity. Nature Immunology 2017, 18, 716-724, 10.1038/ni.3731.
- Claudia Günther; Barbara Kind; Martin A.M. Reijns; Nicole Berndt; Manuel Martínez-Bueno; Christine Wolf; Victoria Tüngler; Osvaldo Chara; Young Ae Lee; Norbert Hubner; et al. Defective removal of ribonucleotides from DNA promotes systemic autoimmunity. Journal of Clinical Investigation 2015, 125, 413-424, 10.1172/JCI78001.
- Zuo; Evan R. Prather; Mykola Stetskiv; Davis E. Garrison; James R. Meade; Timotheus I. Peace; Zhou; Li Zuo; Tingyang Zhou; Inflammaging and Oxidative Stress in Human Diseases: From Molecular Mechanisms to Novel Treatments. International Journal of Molecular Sciences 2019, 20, 4472, 10.3390/ijms20184472.
- Olga A. Sedelnikova; Christophe E. Redon; Jennifer S. Dickey; Asako J. Nakamura; Alexandros G. Georgakilas; William M. Bonner; Role of oxidatively induced DNA lesions in human pathogenesis.. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis 2010, 704, 152-9, 10.1016/j.mrrev.2009.12.005.
- Altieri, F.; Grillo, C.; Maceroni, M.; Chichiarelli, S. DNA damage and repair: From molecular mechanisms to health implications. Antioxid. Redox Signal. 2008, 10, 891–930
- Cˇolak, E.; Ignjatovic ́, S.; Radosavljevic ́, A.; Žoric ́, L. The association of enzymatic and non-enzymatic antioxidant defense parameters with inflammatory markers in patients with exudative form of age-relatedmacular degeneration. J. Clin. Biochem. Nutr. 2017, 60, 100–107.
- Andras Perl; Oxidative stress in the pathology and treatment of systemic lupus erythematosus. Nature Reviews Rheumatology 2013, 9, 674-686, 10.1038/nrrheum.2013.147.
- Doridot, L.; Jeljeli, M.; Chêne, C.; Batteux, F. Implication of oxidative stress in the pathogenesis of systemic sclerosis via inflammation, autoimmunity and fibrosis. Redox Biol. 2019, 101122
- Rosa Vona; Antonello Giovannetti; Lucrezia Gambardella; Walter Malorni; Natella Pietraforte; Elisabetta Straface; Oxidative stress in the pathogenesis of systemic scleroderma: An overview. Journal of Cellular and Molecular Medicine 2018, 22, 3308-3314, 10.1111/jcmm.13630.
- Ozlem Altindag; Mehmet Karakoç; Abdurrahim Kocyigit; Hakim Çelik; Neslihan Soran; Increased DNA damage and oxidative stress in patients with rheumatoid arthritis. Clinical Biochemistry 2007, 40, 167-171, 10.1016/j.clinbiochem.2006.10.006.
- Jessica L. Grieves; Jason M. Fye; Scott Harvey; Jason M. Grayson; Thomas Hollis; Fred W. Perrino; Exonuclease TREX1 degrades double-stranded DNA to prevent spontaneous lupus-like inflammatory disease. Proceedings of the National Academy of Sciences 2015, 112, 5117-5122, 10.1073/pnas.1423804112.
- Philip W. Noble; Sasha Bernatsky; Ann E. Clarke; David A. Isenberg; Rosalind Ramsey-Goldman; James E. Hansen; DNA-damaging autoantibodies and cancer: the lupus butterfly theory. Nature Reviews Rheumatology 2016, 12, 429-434, 10.1038/nrrheum.2016.23.
- Kyung-Jong Lee; Xingwen Dong; Jingsong Wang; Yoshihiko Takeda; William S. Dynan; Identification of human autoantibodies to the DNA ligase IV/XRCC4 complex and mapping of an autoimmune epitope to a potential regulatory region.. The Journal of Immunology 2002, 169, 3413-3421, 10.4049/jimmunol.169.6.3413.
- Victoria L. Fell; Caroline Schild-Poulter; The Ku heterodimer: Function in DNA repair and beyond. Mutation Research/Reviews in Mutation Research 2015, 763, 15-29, 10.1016/j.mrrev.2014.06.002.
- Mahajan, A.; Herrmann, M.; Muñoz, L.E. Clearance Deficiency and Cell Death Pathways: A Model for the Pathogenesis of SLE. Front. Immunol. 2016, 7, 35.
- Jw Park; Sy Moon; Jh Lee; Jk Park; Ds Lee; Kc Jung; Yw Song; Eb Lee; Bone marrow analysis of immune cells and apoptosis in patients with systemic lupus erythematosus. Lupus 2014, 23, 975-985, 10.1177/0961203314531634.
- Paola Sambo; Silvia Svegliati Baroni; Michele Luchetti; Paolo Paroncini; Stefano Dusi; Guido Orlandini; Armando Gabrielli; Oxidative stress in scleroderma: maintenance of scleroderma fibroblast phenotype by the constitutive up-regulation of reactive oxygen species generation through the NADPH oxidase complex pathway.. Arthritis Care & Research 2001, 44, 2653-2664, 10.1002/1529-0131(200111)44:11<2653::aid-art445>3.0.co;2-1.
- Pei-Suen Tsou; Nadine N. Talia; Adam J. Pinney; Ann Kendzicky; Sonsoles Piera-Velázquez; Sergio A. Jimenez; James R. Seibold; Kristine Phillips; Alisa E. Koch; Effect of oxidative stress on protein tyrosine phosphatase 1B in scleroderma dermal fibroblasts.. Arthritis & Rheumatology 2012, 64, 1978-89, 10.1002/art.34336.
- Amélie Servettaz; Claire Goulvestre; Niloufar Kavian; Carole Nicco; Philippe Guilpain; Christiane Chéreau; Vincent Vuiblet; Loïc Guillevin; Luc Mouthon; Bernard Weill; et al. Selective Oxidation of DNA Topoisomerase 1 Induces Systemic Sclerosis in the Mouse. The Journal of Immunology 2009, 182, 5855-5864, 10.4049/jimmunol.0803705.
- Gustavo Martelli Palomino; Carmen L. Bassi; Isabela J. Wastowski; Danilo J. Xavier; Yara M. Lucisano-Valim; Janaina C.O. Crispim; Diane M. Rassi; Joao F. Marques-Neto; Elza T. Sakamoto-Hojo; Philippe Moreau; et al. Patients with Systemic Sclerosis Present Increased DNA Damage Differentially Associated with DNA Repair Gene Polymorphisms. The Journal of Rheumatology 2014, 41, 458-465, 10.3899/jrheum.130376.
- Lan Shao; DNA Damage Response Signals Transduce Stress From Rheumatoid Arthritis Risk Factors Into T Cell Dysfunction. Frontiers in Immunology 2018, 9, 3055, 10.3389/fimmu.2018.03055.
- G. S. Firestein; K. Nguyen; K. R. Aupperle; M. Yeo; D. L. Boyle; N. J. Zvaifler; Apoptosis in rheumatoid arthritis: p53 overexpression in rheumatoid arthritis synovium.. The American Journal of Pathology 1996, 149, 2143-2151, .
- Gary S. Firestein; Fernando Echeverri; Michele Yeo; Nathan J. Zvaifler; Uglas R. Green; Somatic mutations in the p53 tumor suppressor gene in rheumatoid arthritis synovium. Proceedings of the National Academy of Sciences 1997, 94, 10895-10900, 10.1073/pnas.94.20.10895.
- Yuji Yamanishi; David L. Boyle; Sanna Rosengren; Uglas R. Green; Nathan J. Zvaifler; Gary S. Firestein; Regional analysis of p53 mutations in rheumatoid arthritis synovium. Proceedings of the National Academy of Sciences 2002, 99, 10025-10030, 10.1073/pnas.152333199.
- E Šimelyte; D L Boyle; G S Firestein; DNA mismatch repair enzyme expression in synovial tissue. Annals of the Rheumatic Diseases 2004, 63, 1695-1699, 10.1136/ard.2003.017210.
- Martelli-Palomino,G.;Paoliello Paschoalato, A.B.; Crispim, J.C.; Rassi, D.M.; Oliveira, R.D.; Louzada,P.; Lucisano-Valim, Y.M.; Donadi, E.A. DNA damage increase in peripheral neutrophils from patients with rheumatoid arthritis is associated with the disease activity and the presence of shared epitope. Clin. Exp. Rheumatol. 2017, 35, 247–254.
- Vassilis L. Souliotis; Nikolaos I. Vlachogiannis; Maria Pappa; Alexandra Argyriou; Petros P. Sfikakis; DNA damage accumulation, defective chromatin organization and deficient DNA repair capacity in patients with rheumatoid arthritis.. Clinical Immunology 2019, 203, 28-36, 10.1016/j.clim.2019.03.009.
- Mishra, N.; Reilly, C.M.; Brown,D.R.; Ruiz, P.; Gilkeson, G.S. Histone deacetylase inhibitors modulate renal disease in the MRL-lpr / lpr mouse. J. Clin. Invest. 2003, 111, 539–552.
- Nicole L. Regna; Miranda D. Vieson; Xin M. Luo; Cristen B. Chafin; Abdul Gafoor Puthiyaveetil; Sarah E. Hammond; David L. Caudell; Matthew B. Jarpe; Christopher M. Reilly; Specific HDAC6 inhibition by ACY-738 reduces SLE pathogenesis in NZB/W mice. Clinical Immunology 2016, 162, 58-73, 10.1016/j.clim.2015.11.007.
- J. Ren; X. Liao; M. D. Vieson; M. Chen; R. Scott; J. Kazmierczak; X. M. Luo; C. M. Reilly; Selective HDAC6 inhibition decreases early stage of lupus nephritis by down‐regulating both innate and adaptive immune responses. Clinical & Experimental Immunology 2017, 191, 19-31, 10.1111/cei.13046.
- Michaela Waibel; Ailsa J. Christiansen; Margaret L. Hibbs; Jake Shortt; Sarah A. Jones; Ian Simpson; Amanda Light; Kristy O’Donnell; Eric F. Morand; David M. Tarlinton; et al. Manipulation of B-cell responses with histone deacetylase inhibitors. Nature Communications 2015, 6, 6838, 10.1038/ncomms7838.
- Jelena Vojinovic; Nemanja Damjanov; Carmine D'urzo; Antonio Furlan; Gordana Susic; Srdjan Pasic; Nicola Iagaru; Mariana Stefan; Charles A. Dinarello; Safety and efficacy of an oral histone deacetylase inhibitor in systemic-onset juvenile idiopathic arthritis. Arthritis Care & Research 2011, 63, 1452-1458, 10.1002/art.30238.
