Glioblastoma (GB), also known as glioblastoma multiforme, is the most aggressive type of astrocytoma. GB derives both from glial and glioma stem cells and affects glia and astrocytes. Gliomas are heterogeneous neoplasms, classified into grade I to IV according to their malignancy and the presence of specific histological/molecular hallmarks. The higher grade of glioma is known as glioblastoma (GB). Although progress has been made in surgical and radiation treatments, its clinical outcome is still unfavorable. The invasive properties of GB cells and glioma aggressiveness are linked to the reshaping of the cytoskeleton. Recent works suggest that the different susceptibility of GB cells to antitumor immune response is also associated with the extent and function of mitochondria–ER contact sites (MERCs).
- glioblastoma
- glioblastoma invasion
- glioma
- MAMs
- MERCs
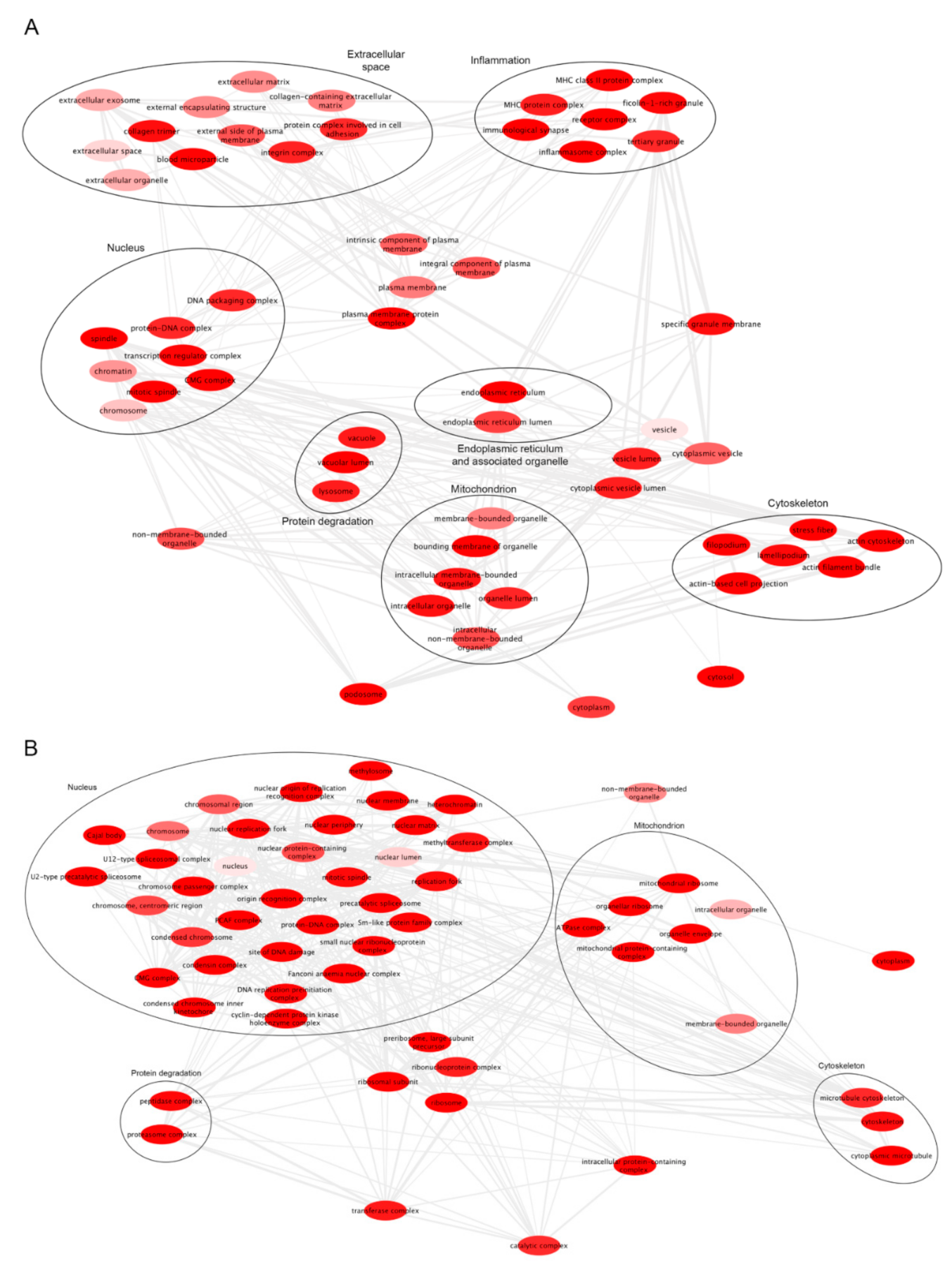
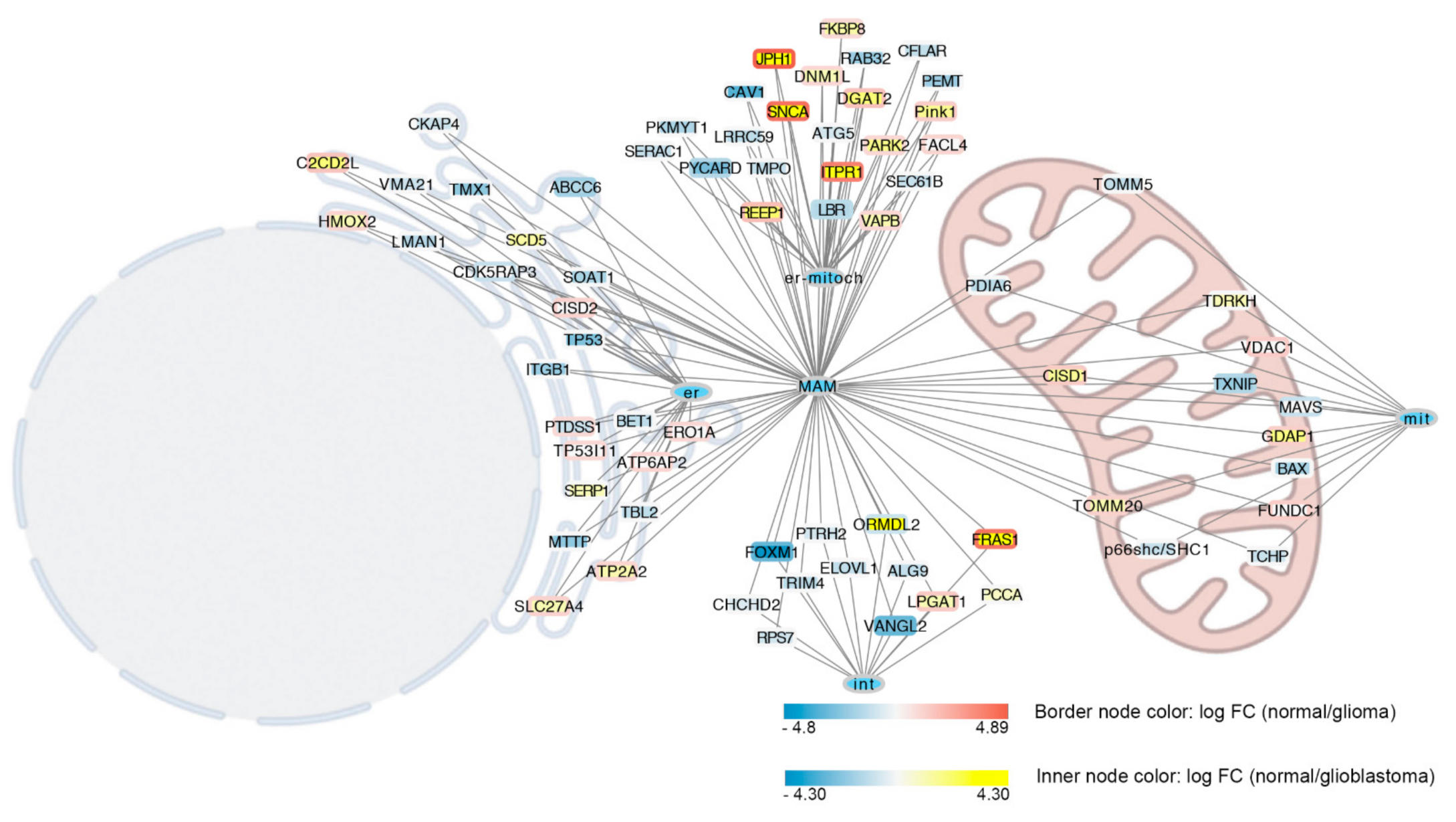
1. MERCs in Glioblastoma
2. MERCs and Microtubules Crosstalk in Glioblastoma
This entry is adapted from the peer-reviewed paper 10.3390/biom12040567
References
- Pinton, P.; Giorgi, C.; Pandolfi, P.P. The Role of PML in the Control of Apoptotic Cell Fate: A New Key Player at ER-Mitochondria Sites. Cell Death Differ. 2011, 18, 1450–1456.
- Molina, J.R.; Hayashi, Y.; Stephens, C.; Georgescu, M.M. Invasive Glioblastoma Cells Acquire Stemness and Increased Akt Activation. Neoplasia 2010, 12, 453–463.
- Bassoy, E.Y.; Kasahara, A.; Chiusolo, V.; Jacquemin, G.; Boydell, E.; Zamorano, S.; Riccadonna, C.; Pellegatta, S.; Hulo, N.; Dutoit, V.; et al. ER–Mitochondria Contacts Control Surface Glycan Expression and Sensitivity to Killer Lymphocytes in Glioma Stem-like Cells. EMBO J. 2017, 36, 1493–1512.
- Arismendi-Morillo, G.J.; Castellano-Ramirez, A.V. Ultrastructural Mitochondrial Pathology in Human Astrocytic Tumors: Potentials Implications pro-Therapeutics Strategies. J. Electron. Microsc. 2008, 57, 33–39.
- Vlashi, E.; Lagadec, C.; Vergnes, L.; Matsutani, T.; Masui, K.; Poulou, M.; Popescu, R.; della Donna, L.; Evers, P.; Dekmezian, C.; et al. Metabolic State of Glioma Stem Cells and Nontumorigenic Cells. Proc. Natl. Acad. Sci. USA 2011, 108, 16062–16067.
- Arismendi-Morillo, G.; Castellano-Ramírez, A.; Seyfried, T.N. Functional and Therapeutic Implications of Mitochondrial Network and Mitochondria-Associated Membranes: The Glioma’s Case. Glioma-Contemp. Diagn. Ther. Approaches 2019.
- Giacomello, M.; Drago, I.; Pizzo, P.; Pozzan, T. Mitochondrial Ca2+ as a Key Regulator of Cell Life and Death. Cell Death Differ. 2007, 14, 1267–1274.
- Moser, J.J.; Fritzler, M.J.; Rattner, J.B. Ultrastructural Characterization of Primary Cilia in Pathologically Characterized Human Glioblastoma Multiforme (GBM) Tumors. BMC Clin. Pathol. 2014, 14, 40.
- Moser, J.J.; Fritzler, M.J.; Rattner, J.B. Primary Ciliogenesis Defects Are Associated with Human Astrocytoma/Glioblastoma Cells. BMC Cancer 2009, 9, 448.
- Boehning, D.; Patterson, R.L.; Sedaghat, L.; Glebova, N.O.; Kurosaki, T.; Snyder, S.H. Cytochrome c Binds to Inositol (1,4,5) Trisphosphate Receptors, Amplifying Calcium-Dependent Apoptosis. Nat. Cell Biol. 2003, 5, 1051–1061.
- Carré, M.; André, N.; Carles, G.; Borghi, H.; Brichese, L.; Briand, C.; Braguer, D.; Medicine, U.F.R.; La, U.O. Tubulin Is an Inherent Component of Mitochondrial Membranes That Interacts with the Voltage-Dependent Anion Channel. J. Biol. Chem. 2002, 277, 33664–33669.
- Puurand, M.; Tepp, K.; Timohhina, N.; Aid, J.; Shevchuk, I.; Chekulayev, V.; Kaambre, T. Tubulin ΒII and ΒIII Isoforms as the Regulators of VDAC Channel Permeability in Health and Disease. Cells 2019, 8, 239.
- Maldonado, E.N.; Sheldon, K.L.; Dehart, D.N.; Patnaik, J.; Manevich, Y.; Townsend, D.M.; Bezrukov, S.M.; Rostovtseva, T.K.; Lemasters, J.J. Voltage-Dependent Anion Channels Modulate Mitochondrial Metabolism in Cancer Cells: Regulation by Free Tubulin and Erastin. J. Biol. Chem. 2013, 288, 11920–11929.
- Strickland, M.; Stoll, E.A. Metabolic Reprogramming in Glioma. Front. Cell Dev. Biol. 2017, 5, 43.
- Mucignat-Caretta, C.; Cavaggioni, A.; Redaelli, M.; Malatesta, M.; Zancanaro, C.; Caretta, A. Selective Distribution of Protein Kinase A Regulatory Subunit Rllα in Rodent Gliomas. Neuro-Oncology 2008, 10, 958–967.
- Zhao, P.; Li, Q.; Shi, Z.; Li, C.; Wang, L.; Liu, X.; Jiang, C.; Qian, X.; You, Y.; Liu, N.; et al. GSK-3β Regulates Tumor Growth and Angiogenesis in Human Glioma Cells Theseauthorshavecontributedequallytothiswork. Oncotarget 2021, 6, 31901–31915.
- Collet, B.; Guitton, N.; Saïkali, S.; Avril, T.; Pineau, C.; Hamlat, A.; Mosser, J.; Quillien, V. Differential Analysis of Glioblastoma Multiforme Proteome by a 2D-DIGE Approach. Proteome Sci. 2011, 9, 16.
- Cui, Y.; Li, J.; Weng, L.; Wirbisky, S.E.; Freeman, J.L.; Liu, J.; Liu, Q.; Yuan, X.; Irudayaraj, J. Regulatory Landscape and Clinical Implication of MBD3 in Human Malignant Glioma. Oncotarget 2016, 7, 81698–81714.
- Emig, D.; Salomonis, N.; Baumbach, J.; Lengauer, T.; Conklin, B.R.; Albrecht, M. AltAnalyze and DomainGraph: Analyzing and Visualizing Exon Expression Data. Nucleic Acids Res. 2010, 38, 755–762.
- Alessio, E.; Buson, L.; Chemello, F.; Peggion, C.; Grespi, F.; Martini, P.; Massimino, M.L.; Pacchioni, B.; Millino, C.; Romualdi, C.; et al. Single Cell Analysis Reveals the Involvement of the Long Non-Coding RNA Pvt1 in the Modulation of Muscle Atrophy and Mitochondrial Network. Nucleic Acids Res. 2019, 47, 1653–1670.
- Pagliari, M.; Munari, F.; Toffoletto, M.; Lonardi, S.; Chemello, F.; Codolo, G.; Millino, C.; Della Bella, C.; Pacchioni, B.; Vermi, W.; et al. Helicobacter Pylori Affects the Antigen Presentation Activity of Macrophages Modulating the Expression of the Immune Receptor CD300E through MiR-4270. Front. Immunol. 2017, 8, 1288.
- Howe, E.; Holton, K.; Nair, S.; Schlauch, D.; Sinha, R.; Quackenbush, J. MeV: MultiExperiment Viewer. In Biomedical Informatics for Cancer Research; Springer: Berlin/Heidelberg, Germany, 2010; pp. 267–277. ISBN 9781441957122.
- Mi, H.; Thomas, P. PANTHER Pathway: An Ontology-Based Pathway Database Coupled with Data Analysis Tools. Methods Mol. Biol. 2009, 563, 123–140.
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. Revigo Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS ONE 2011, 6, e21800.
- Schweizer, A.; Ericsson, M.; Bachi, T.; Griffiths, G.; Hauri, H.P. Characterization of a Novel 63 KDa Membrane Protein. Implications for the Organization of the ER-to-Golgi Pathway. J. Cell Sci. 1993, 104, 671–683.
- Harada, T.; Sada, R.; Osugi, Y.; Matsumoto, S.; Matsuda, T.; Hayashi-Nishino, M.; Nagai, T.; Harada, A.; Kikuchi, A. Palmitoylated CKAP4 Regulates Mitochondrial Functions through an Interaction with VDAC2 at ER-Mitochondria Contact Sites. J. Cell Sci. 2020, 133, jcs249045.
- Kikuchi, A.; Fumoto, K.; Kimura, H. The Dickkopf1-Cytoskeleton-Associated Protein 4 Axis Creates a Novel Signalling Pathway and May Represent a Molecular Target for Cancer Therapy. Br. J. Pharmacol. 2017, 174, 4651–4665.
- Noda, C.; Kimura, H.; Arasaki, K.; Matsushita, M.; Yamamoto, A.; Wakana, Y.; Inoue, H.; Tagaya, M. Valosin-Containing Protein-Interacting Membrane Protein (VIMP) Links the Endoplasmic Reticulum with Microtubules in Concert with Cytoskeleton-Linking Membrane Protein (CLIMP)-63. J. Biol. Chem. 2014, 289, 24304–24313.
