Biosensors have globally been considered as biomedical diagnostic tools required in abundant areas including the development of diseases, detection of viruses, diagnosing ecological pollution, food monitoring, and a wide range of other diagnostic and therapeutic biomedical research. Recently, the broadly emerging and promising technique of plasmonic resonance has proven to provide label-free and highly sensitive real-time analysis when used in biosensing applications.
- biosensors
- plasmonics
- surface plasmon resonance
- lab-on-chip
1. Introduction
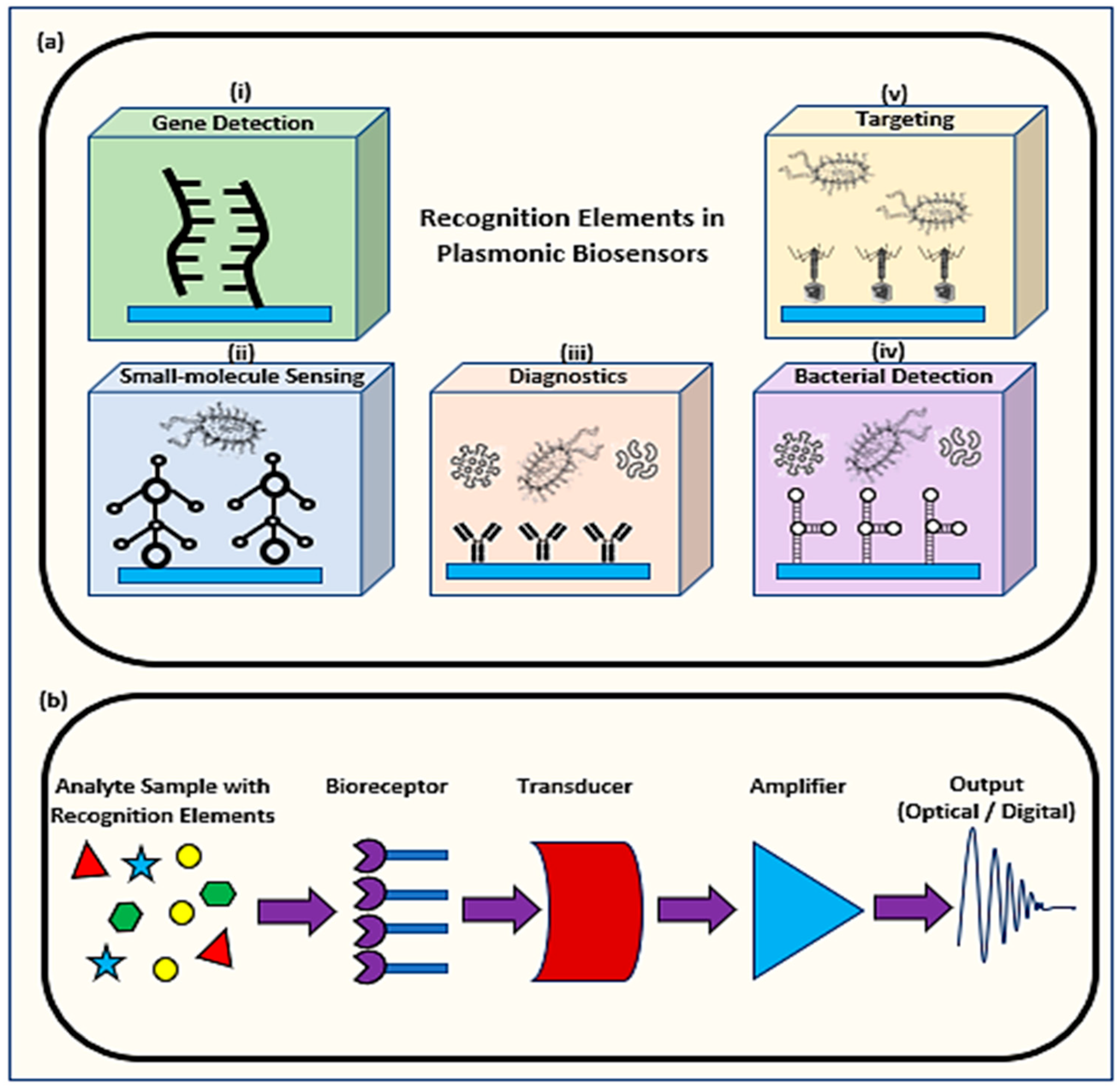
1.1. Mechanism of Plasmonic Biosensors
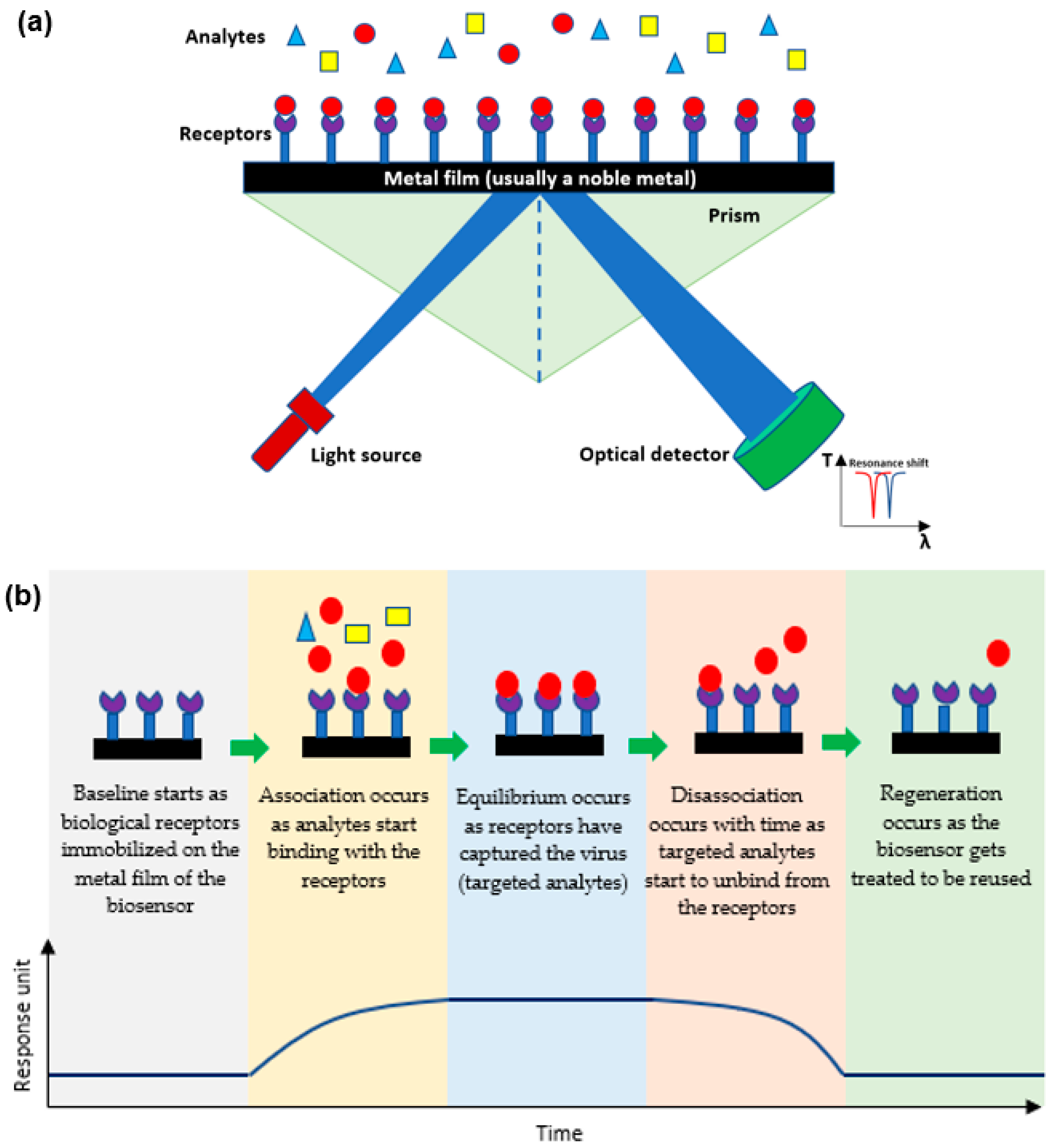
1.2. Determining the Efficiency of a Plasmonic Biosensor
2. Applications of Plasmonic Optical Biosensors
2.1. The Use of Plasmonic Biosensors for Viral Detection
|
Current Methods for Virus Detection |
Advantages |
Limitations |
Refs. |
|---|---|---|---|
|
Immunofluorescence Assays |
Numerous, simultaneous samples can be analyzed and stored for some time. |
Fluorescent molecules bound to primary antibody is limited. Low sensitivity may result in false negatives. |
[46] |
|
Hemagglutination Assays |
Low-cost instruments. Results within hours. Has standardization as it is recognized in labs worldwide. |
Little specificity. Requires trained personnel. Analysis needed by qualified individuals. Difficult inter-laboratory comparison of results due to the several controlled variables. |
[47] |
|
Viral Plaque Assay |
Available in most labs. Rapid results. |
Absence of standardization. Involves costly repeat runs for accurate results. |
[48] |
|
Viral Flow Cytometry |
Rapid results. Numerous cells analyzed instantly. |
Requires highly trained personnel. Requires ongoing maintenance by service engineers. |
[49] |
|
ELISA |
Accurate/fast results. Very sensitive process. Easily automated. |
Expensive preparation method. Requires trained personnel. |
[40] |
|
CT |
Combined assessment. Short acquisition time. |
Expensive preparation method. Requires trained personnel. Exposure to gamma rays. |
[41] |
|
NAAT |
Very sensitive process. Accurate and reliable |
Requires trained personnel. Expensive detection kits. Time-consuming (2–3 days). False-positive cases. |
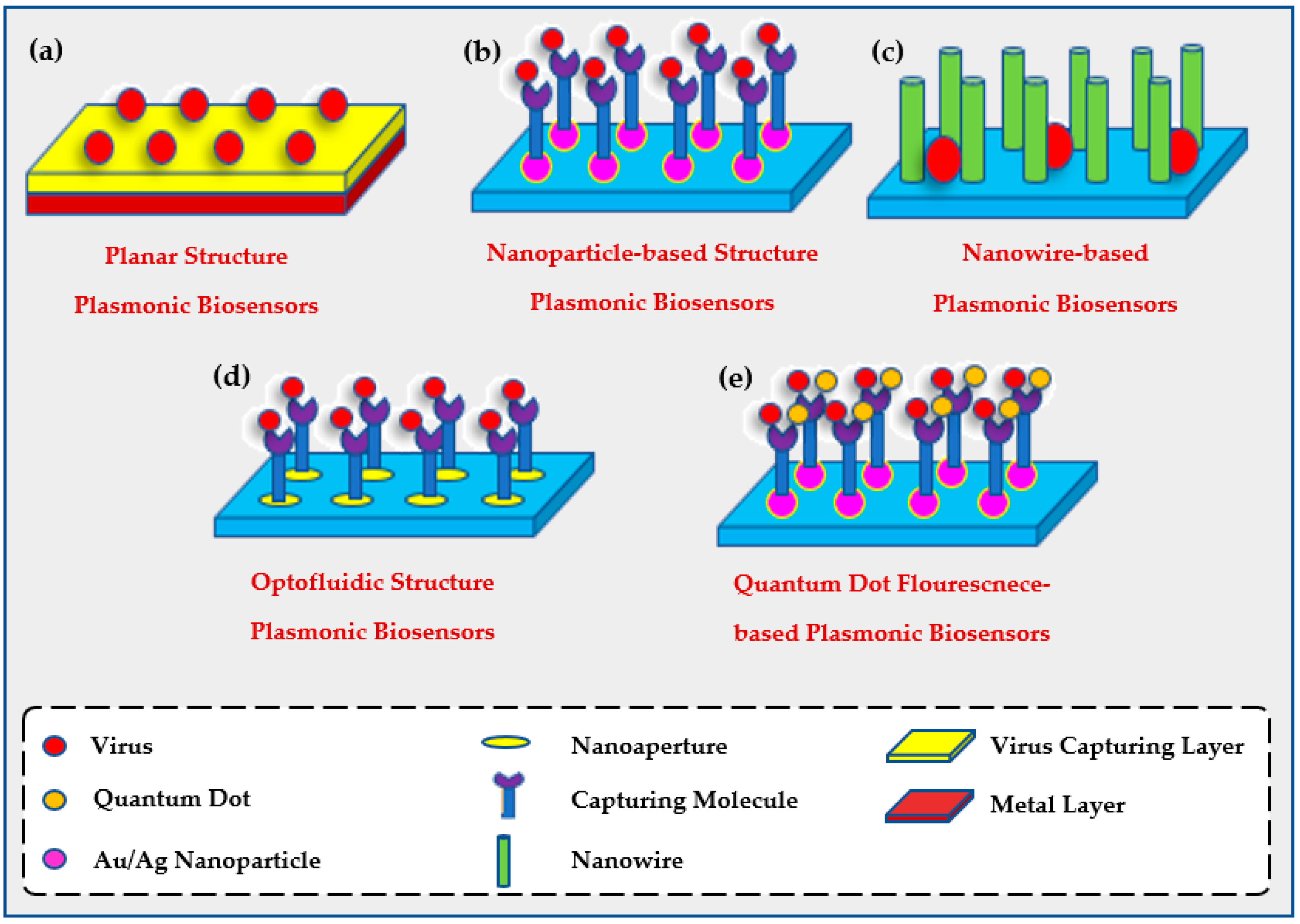
2.2. The Use of Plasmonic Biosensors for Environmental Evaluation
2.3. The Use of Plasmonic Biosensors for Food Analysis
|
Component |
Other Methods |
SPR |
Refs. |
|---|---|---|---|
|
Heavy metals |
Atomic Absorption Spectroscopy
Inductively Coupled Plasma Mass Spectrometry
X-ray Fluorescence Spectrometry
|
|
|
|
Food Allergens |
ELISA
|
|
|
|
Citrinin (Mycotoxin) |
HPLC and LC-MS
|
|
|
|
Pesticides |
LC-MS/MS
|
|
|
|
β-Lactoglobulin |
ELISA and LC-MS
|
|
|
|
Tetrodotoxin (Fish toxin) |
LC-MS/MS, ELISA, and HPLC
|
|
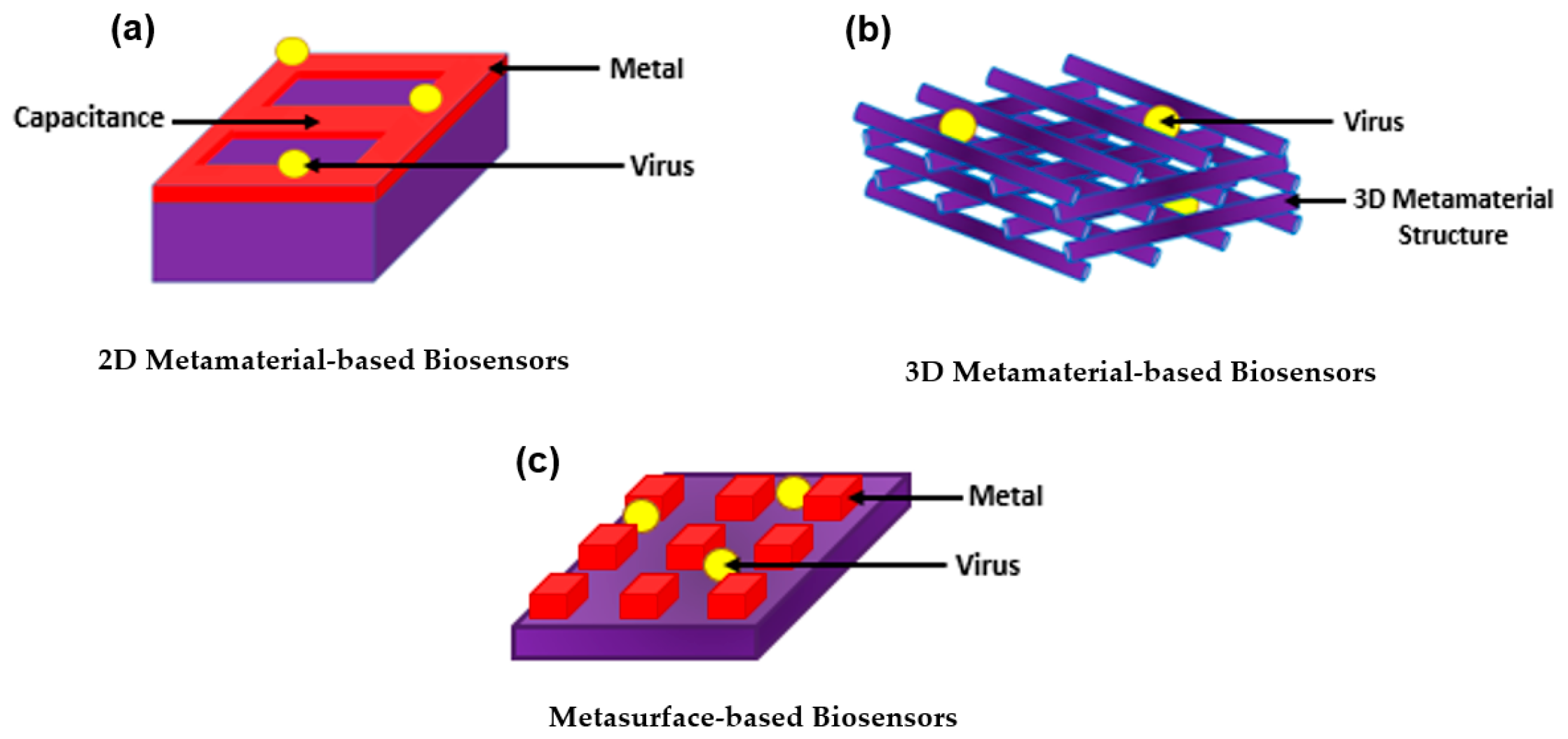
3. Introducing Metamaterials to Plasmonic Biosensors
4. Lab-on-a-Chip (LoC) for Plasmonic Biosensors
This entry is adapted from the peer-reviewed paper 10.3390/biology11050621
References
- Villalonga, A.; Pérez-Calabuig, A.M.; Villalonga, R. Electrochemical biosensors based on nucleic acid aptamers. Anal. Bioanal. Chem. 2020, 412, 55–72.
- Jadhav, D.A.; Chendake, A.D.; Ghosal, D.; Mathuriya, A.S.; Kumar, S.S.; Pandit, S. Advanced microbial fuel cell for biosensor applications to detect quality parameters of pollutants. In Bioremediation, Nutrients, and Other Valuable Product Recovery; Singh, L., Mahapatra, D.M., Thakur, S., Eds.; Elsevier: Amsterdam, The Netherlands, 2021; pp. 125–139.
- Bollella, P.; Katz, E. Biosensors—Recent Advances and Future Challenges. Sensors 2020, 20, 6645.
- Nguyen, H.H.; Lee, S.; Lee, U.; Fermin, C.; Kim, M. Immobilized Enzymes in Biosensor Applications. Materials 2019, 12, 121.
- Mauriz, E.; Lechuga, L.M. Plasmonic Biosensors for Single-Molecule Biomedical Analysis. Biosensors 2021, 11, 123.
- Solhi, E.; Hasanzadeh, M. Critical role of biosensing on the efficient monitoring of cancer proteins/biomarkers using label-free aptamer based bioassay. Biomed. Pharmacother. 2020, 132, 110849.
- Sezgintürk, M.K. Introduction to commercial biosensors. In Commercial Biosensors and Their Applications; Sezgintürk, M.K., Ed.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 1–28.
- Mehrotra, P. Biosensors and their applications—A review. J. Oral Biol. Craniofac. Res. 2016, 6, 153–159.
- Kim, J.; Campbell, A.S.; de Ávila, B.E.-F.; Wang, J. Wearable biosensors for healthcare monitoring. Nat. Biotechnol. 2019, 37, 389–406.
- Wu, K.; Saha, R.; Su, D.; Krishna, V.D.; Liu, J.; Cheeran, M.C.-J.; Wang, J.-P. Magnetic-Nanosensor-Based Virus and Pathogen Detection Strategies before and during COVID-19. ACS Appl. Nano Mater. 2020, 3, 9560–9580.
- Zhao, V.X.T.; Wong, T.I.; Zheng, X.T.; Tan, Y.N.; Zhou, X. Colorimetric biosensors for point-of-care virus detections. Mater. Sci. Energy Technol. 2020, 3, 237–249.
- Kaya, S.I.; Karadurmus, L.; Ozcelikay, G.; Bakirhan, N.K.; Ozkan, S.A. Electrochemical virus detections with nanobiosensors. Nanosens. Smart Cities 2020, 303–326.
- Antiochia, R. Paper-Based Biosensors: Frontiers in Point-of-Care Detection of COVID-19 Disease. Biosensors 2021, 11, 110.
- Maddali, H.; Miles, C.E.; Kohn, J.; O’Carroll, D.M. Optical Biosensors for Virus Detection: Prospects for SARS-CoV-2/COVID-19. Chembiochem 2021, 22, 1176–1189.
- Zanchetta, G.; Lanfranco, R.; Giavazzi, F.; Bellini, T.; Buscaglia, M. Emerging applications of label-free optical biosensors. Nanophotonics 2017, 6, 627–645.
- Liu, Y.; Zhang, X. Microfluidics-Based Plasmonic Biosensing System Based on Patterned Plasmonic Nanostructure Arrays. Micromachines 2021, 12, 826.
- Liu, L.; Ye, K.; Jia, Z.; Xue, T.; Nie, A.; Xiang, J.; Mu, C.; Wang, B.; Wen, F.; Zhai, K.; et al. High-sensitivity and versatile plasmonic biosensor based on grain boundaries in polycrystalline 1L WS2 films. Biosens. Bioelectron. 2021, 194, 113596.
- Li, X.; Soler, M.; Özdemir, C.I.; Belushkin, A.; Yesilköy, F.; Altug, H. Plasmonic nanohole array biosensor for label-free and real-time analysis of live cell secretion. Lab Chip 2017, 17, 2208–2217.
- Armstrong, R.E.; Horáček, M.; Zijlstra, P. Plasmonic Assemblies for Real-Time Single-Molecule Biosensing. Small 2020, 16, 2003934.
- Minopoli, A.; Della Ventura, B.; Lenyk, B.; Gentile, F.; Tanner, J.A.; Offenhäusser, A.; Mayer, D.; Velotta, R. Ultrasensitive antibody-aptamer plasmonic biosensor for malaria biomarker detection in whole blood. Nat. Commun. 2020, 11, 6134.
- Mondal, B.; Zeng, S. Recent Advances in Surface Plasmon Resonance for Biosensing Applications and Future Prospects. 2020. Available online: https://hal-unilim.archives-ouvertes.fr/hal-02915657 (accessed on 18 March 2022).
- Duan, Q.; Liu, Y.; Chang, S.; Chen, H.; Chen, J. Surface Plasmonic Sensors: Sensing Mechanism and Recent Applications. Sensors 2021, 21, 5262.
- Hill, R.T. Plasmonic Biosensors. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2015, 7, 152–168.
- Miyazaki, C.; Shimizu, F.; Ferreira, M. Surface Plasmon Resonance (SPR) for Sensors and Biosensors. In Nanocharacterization Techniques; William Andrew Publishing: Norwich, NY, USA, 2017; pp. 183–200.
- Chlebus, R.; Chylek, J.; Ciprian, D.; Hlubina, P. Surface Plasmon Resonance Based Measurement of the Dielectric Function of a Thin Metal Film. Sensors 2018, 18, 3693.
- Kravets, V.G.; Wu, F.; Yu, T.; Grigorenko, A.N. Metal-Dielectric-Graphene Hybrid Heterostructures with Enhanced Surface Plasmon Resonance Sensitivity Based on Amplitude and Phase Measurements. Plasmonics 2022.
- Zakaria, R.; Zainuddin, N.A.M.; Leong, T.C.; Rosli, R.; Rusdi, M.F.; Harun, S.W.; Sadegh Amiri, I. Investigation of Surface Plasmon Resonance (SPR) in MoS2- and WS2-Protected Titanium Side-Polished Optical Fiber as a Humidity Sensor. Micromachines 2019, 10, 465.
- Derkachova, A.; Kolwas, K.; Demchenko, I. Dielectric Function for Gold in Plasmonics Applications: Size Dependence of Plasmon Resonance Frequencies and Damping Rates for Nanospheres. Plasmonics 2016, 11, 941–951.
- Raza, S.; Stenger, N.; Kadkhodazadeh, S.; Fischer, S.V.; Kostesha, N.; Jauho, A.-P.; Burrows, A.; Wubs, M.; Mortensen, N.A. Blueshift of the surface plasmon resonance in silver nanoparticles studied with EELS. Nanophotonics 2013, 2, 131–138.
- Zhu, X.; Gao, T. Spectrometry. In Nano-Inspired Biosensors for Protein Assay with Clinical Applications; Li, G., Ed.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 237–264.
- Hassan, M.M.; Sium, F.S.; Islam, F.; Choudhury, S.M. A review on plasmonic and metamaterial based biosensing platforms for virus detection. Sens. Bio-Sens. Res. 2021, 33, 100429.
- Zhou, X.; Chen, K.; Li, L.; Peng, W.; Yu, Q. Angle modulated surface plasmon resonance spectrometer for refractive index sensing with enhanced detection resolution. Opt. Commun. 2017, 382, 610–614.
- Xu, K.; Zhou, R.; Takei, K.; Hong, M. Toward Flexible Surface-Enhanced Raman Scattering (SERS) Sensors for Point-of-Care Diagnostics. Adv. Sci. 2019, 6, 1900925.
- Park, S.-G.; Mun, C.; Xiao, X.; Braun, A.; Kim, S.; Giannini, V.; Maier, S.A.; Kim, D.-H. Surface Energy-Controlled SERS Substrates for Molecular Concentration at Plasmonic Nanogaps. Adv. Funct. Mater. 2017, 27, 1703376.
- Elsayed, M.; Gouda, A.; Ismail, Y.; Swillam, M. Silicon-Based SERS Substrates Fabricated by Electroless Etching. J. Lightwave Technol. 2017, 35, 3075–3081.
- EP17A2|Evaluation of Detection Capability for Clinical Laboratory Measurement Procedures, 2nd ed. Available online: https://www.clsi.orghttps://clsi.org/standards/products/method-evaluation/documents/ep17/ (accessed on 18 March 2022).
- WHO Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int (accessed on 22 July 2021).
- Louten, J. Detection and Diagnosis of Viral Infections. Essent. Hum. Virol. 2016, 111–132.
- Burrell, C.J.; Howard, C.R.; Murphy, F.A. Laboratory Diagnosis of Virus Diseases. Fenner White’s Med. Virol. 2017, 135–154.
- Zhao, X.; Lin, C.-W.; Wang, J.; Oh, D.H. Advances in Rapid Detection Methods for Foodborne Pathogens. J. Microbiol. Biotechnol. 2014, 24, 297–312.
- Bağcı, U.; Bray, M.; Caban, J.; Yao, J.; Mollura, D.J. Computer-Assisted Detection of Infectious Lung Diseases: A Review. Comput. Med. Imaging Graph. 2012, 36, 72–84.
- Kumar, P. Methods for Rapid Virus Identification and Quantification. Mater. Methods 2013, 3, 13070.
- Yamamoto, K.; Ohmagari, N. Microbiological Testing for Coronavirus Disease 2019. JMA J. 2021, 4, 67–75.
- Bruce, E.A.; Mills, M.G.; Sampoleo, R.; Perchetti, G.A.; Huang, M.-L.; Despres, H.W.; Schmidt, M.M.; Roychoudhury, P.; Shirley, D.J.; Jerome, K.R.; et al. Predicting infectivity: Comparing four PCR-based assays to detect culturable SARS-CoV-2 in clinical samples. EMBO Mol. Med. 2022, 14, e15290.
- Ahmed, W.; Simpson, S.L.; Bertsch, P.M.; Bibby, K.; Bivins, A.; Blackall, L.L.; Bofill-Mas, S.; Bosch, A.; Brandão, J.; Choi, P.M.; et al. Minimizing errors in RT-PCR detection and quantification of SARS-CoV-2 RNA for wastewater surveillance. Sci. Total Environ. 2022, 805, 149877.
- Toya, M.; Iida, Y.; Sugimoto, A. Imaging of Mitotic Spindle Dynamics in Caenorhabditis elegans Embryos. In Methods in Cell Biology; Cassimeris, L., Tran, P., Eds.; Microtubules: In vivo; Academic Press: Cambridge, MA, USA, 2010; Volume 97, pp. 359–372.
- Noah, D.L.; Hill, H.; Hines, D.; White, E.L.; Wolff, M.C. Qualification of the Hemagglutination Inhibition Assay in Support of Pandemic Influenza Vaccine Licensure. Clin. Vaccine Immunol. 2009, 16, 558–566.
- Mendoza, E.J.; Manguiat, K.; Wood, H.; Drebot, M. Two Detailed Plaque Assay Protocols for the Quantification of Infectious SARS-CoV-2. Curr. Protoc. Microbiol. 2020, 57, ecpmc105.
- Cossarizza, A.; Chang, H.-D.; Radbruch, A.; Acs, A.; Adam, D.; Adam-Klages, S.; Agace, W.W.; Aghaeepour, N.; Akdis, M.; Allez, M.; et al. Guidelines for the use of flow cytometry and cell sorting in immunological studies (second edition). Eur. J. Immunol. 2019, 49, 1457–1973.
- Tymm, C.; Zhou, J.; Tadimety, A.; Burklund, A.; Zhang, J.X.J. Scalable COVID-19 Detection Enabled by Lab-on-Chip Biosensors. Cell. Mol. Bioeng. 2020, 13, 313–329.
- Rodriguez-Mozaz, S.; Marco, M.-P.; de Alda, M.J.L.; Barceló, D. Biosensors for environmental applications: Future development trends. Pure Appl. Chem. 2004, 76, 723–752.
- Rodriguez-Mozaz, S.; de Alda, M.J.L.; Marco, M.-P.; Barceló, D. Biosensors for environmental monitoring A global perspective. Talanta 2005, 65, 291–297.
- Rogers, K.R. Recent advances in biosensor techniques for environmental monitoring. Anal. Chim. Acta 2006, 568, 222–231.
- Rogers, K.R.; Gerlach, C.L. Peer Reviewed: Environmental Biosensors: A Status Report. Environ. Sci. Technol. 1996, 30, 486A–491A.
- Ju, H.; Kandimalla, V.B. Biosensors for pesticides. In Electrochemical Sensors, Biosensors and Their Biomedical Applications; Zhang, X., Ju, H., Wang, J., Eds.; Academic Press: San Diego, CA, USA, 2008; pp. 31–56.
- Suryan, S.K. Biosensors: A Tool for Environmental Monitoring and Analysis. In Advances in Environmental Biotechnology; Kumar, R., Sharma, A.K., Ahluwalia, S.S., Eds.; Springer: Singapore, 2017; pp. 265–288.
- Gieva, E.; Nikolov, G.; Nikolova, B. Biosensors for Environmental Monitoring. In Proceedings of the Conference on Challenges in Higher Education & Research, Proceedings of the 12th Conference, Sozopol, Bulgaria, 17–19 November 2014; Volume 12.
- Food Safety. Available online: https://www.who.int/news-room/fact-sheets/detail/food-safety (accessed on 30 December 2021).
- Ravindran, N.; Kumar, S.; Yashini, M.; Rajeshwari, S.; Mamathi, C.A.; Nirmal Thirunavookarasu, S.N.; Sunil, C.K. Recent advances in Surface Plasmon Resonance (SPR) biosensors for food analysis: A review. Crit. Rev. Food Sci. Nutr. 2021, 1–23.
- Narsaiah, K.; Jha, S.N.; Bhardwaj, R.; Sharma, R.; Kumar, R. Optical biosensors for food quality and safety assurance—A review. J. Food Sci. Technol. 2012, 49, 383.
- Wing Fen, Y.; Mahmood Mat Yunus, W. Surface plasmon resonance spectroscopy as an alternative for sensing heavy metal ions: A review. Sens. Rev. 2013, 33, 305–314.
- Verma, R.; Gupta, B.D. Detection of heavy metal ions in contaminated water by surface plasmon resonance based optical fibre sensor using conducting polymer and chitosan. Food Chem. 2015, 166, 568–575.
- Ashley, J.; Shahbazi, M.-A.; Kant, K.; Chidambara, V.A.; Wolff, A.; Bang, D.D.; Sun, Y. Molecularly imprinted polymers for sample preparation and biosensing in food analysis: Progress and perspectives. Biosens. Bioelectron. 2017, 91, 606–615.
- Atar, N.; Eren, T.; Yola, M.L. A molecular imprinted SPR biosensor for sensitive determination of citrinin in red yeast rice. Food Chem. 2015, 184, 7–11.
- Zhou, J.; Qi, Q.; Wang, C.; Qian, Y.; Liu, G.; Wang, Y.; Fu, L. Surface plasmon resonance (SPR) biosensors for food allergen detection in food matrices. Biosens. Bioelectron. 2019, 142, 111449.
- Çakır, O.; Baysal, Z. Pesticide analysis with molecularly imprinted nanofilms using surface plasmon resonance sensor and LC-MS/MS: Comparative study for environmental water samples. Sens. Actuators B Chem. 2019, 297, 126764.
- D’Aurelio, R.; Ashley, J.; Rodgers, T.L.; Trinh, L.; Temblay, J.; Pleasants, M.; Tothill, I.E. Development of a NanoMIPs-SPR-Based Sensor for β-Lactoglobulin Detection. Chemosensors 2020, 8, 94.
- Ashley, J.; D’Aurelio, R.; Piekarska, M.; Temblay, J.; Pleasants, M.; Trinh, L.; Rodgers, T.L.; Tothill, I.E. Development of a β-Lactoglobulin Sensor Based on SPR for Milk Allergens Detection. Biosensors 2018, 8, 32.
- Yakes, B.J.; Deeds, J.; White, K.; Degrasse, S.L. Evaluation of surface plasmon resonance biosensors for detection of tetrodotoxin in food matrices and comparison to analytical methods. J. Agric. Food Chem. 2011, 59, 839–846.
- Hodnik, V.; Anderluh, G. Toxin detection by surface plasmon resonance. Sensors 2009, 9, 1339–1354.
- Chalyan, T.; Potrich, C.; Schreuder, E.; Falke, F.; Pasquardini, L.; Pederzolli, C.; Heideman, R.; Pavesi, L. AFM1 Detection in Milk by Fab’ Functionalized Si3N4 Asymmetric Mach-Zehnder Interferometric Biosensors. Toxins 2019, 11, 409.
- Smith, D.R.; Padilla, W.J.; Vier, D.C.; Nemat-Nasser, S.C.; Schultz, S. Composite Medium with Simultaneously Negative Permeability and Permittivity. Phys. Rev. Lett. 2000, 84, 4184–4187.
- Beruete, M.; Jáuregui-López, I. Terahertz Sensing Based on Metasurfaces. Adv. Opt. Mater. 2020, 8, 1900721.
- Chen, T.; Li, S.; Sun, H. Metamaterials application in sensing. Sensors 2012, 12, 2742–2765.
- Zhou, R.; Wang, C.; Xu, W.; Xie, L. Biological applications of terahertz technology based on nanomaterials and nanostructures. Nanoscale 2019, 11, 3445–3457.
- Ahmadivand, A.; Gerislioglu, B.; Tomitaka, A.; Manickam, P.; Kaushik, A.; Bhansali, S.; Nair, M.; Pala, N. Extreme sensitive metasensor for targeted biomarkers identification using colloidal nanoparticles-integrated plasmonic unit cells. Biomed. Opt. Express 2018, 9, 373–386.
- Lee, D.-K.; Kang, J.-H.; Kwon, J.; Lee, J.-S.; Lee, S.; Woo, D.H.; Kim, J.H.; Song, C.-S.; Park, Q.-H.; Seo, M. Nano metamaterials for ultrasensitive Terahertz biosensing. Sci. Rep. 2017, 7, 8146.
- Ahmed, R.; Ozen, M.O.; Karaaslan, M.G.; Prator, C.A.; Thanh, C.; Kumar, S.; Torres, L.; Iyer, N.; Munter, S.; Southern, S.; et al. Tunable Fano-Resonant Metasurfaces on a Disposable Plastic-Template for Multimodal and Multiplex Biosensing. Adv. Mater. 2020, 32, e1907160.
- Park, S.J.; Cha, S.H.; Shin, G.A.; Ahn, Y.H. Sensing viruses using terahertz nano-gap metamaterials. Biomed. Opt. Express 2017, 8, 3551–3558.
- Sreekanth, K.V.; El Kabbash, M.; Alapan, Y.; Ilker, E.I.; Hinczewski, M.; Gurkan, U.A.; Strangi, G. Hyperbolic metamaterials-based plasmonic biosensor for fluid biopsy with single molecule sensitivity. EPJ Appl. Metamater. 2017, 4, 1.
- Aristov, A.I.; Manousidaki, M.; Danilov, A.; Terzaki, K.; Fotakis, C.; Farsari, M.; Kabashin, A.V. 3D plasmonic crystal metamaterials for ultra-sensitive biosensing. Sci. Rep. 2016, 6, 25380.
- Cheng, D.; He, X.; Huang, X.; Zhang, B.; Liu, G.; Shu, G.; Fang, C.; Wang, J.; Luo, Y. Terahertz biosensing metamaterial absorber for virus detection based on spoof surface plasmon polaritons. Int. J. RF Microw. Comput.-Aided Eng. 2018, 28, e21448.
- Ahmadivand, A.; Gerislioglu, B.; Manickam, P.; Kaushik, A.; Bhansali, S.; Nair, M.; Pala, N. Rapid Detection of Infectious Envelope Proteins by Magnetoplasmonic Toroidal Metasensors. ACS Sens. 2017, 2, 1359–1368.
- Alam, M.M.; Malik, H.; Khan, M.I.; Pardy, T.; Kuusik, A.; Le Moullec, Y. A Survey on the Roles of Communication Technologies in IoT-Based Personalized Healthcare Applications. IEEE Access 2018, 6, 36611–36631.
- Kotb, R.; Ismail, Y.; Swillam, M. Integrated metal-insulator-metal plasmonic nano resonator: An analytical approach. Prog. Electromagn. Res. Lett. 2013, 43, 83–94.
- El-Zohary, S.E.; Azzazi, A.A.; Okamoto, H.; Okamoto, T.; Haraguchi, M.; Swillam, M.A. Resonance-based integrated plasmonic nanosensor for lab-on-chip applications. JNP 2013, 7, 073077.
- Swillam, M.; Helmy, A. Analysis and applications of 3D rectangular metallic waveguides. Opt. Express 2010, 18, 19831–19843.
- Sherif, S.M.; Zografopoulos, D.C.; Shahada, L.A.; Beccherelli, R.; Swillam, M. Integrated plasmonic refractometric sensor using Fano resonance. J. Phys. D Appl. Phys. 2017, 50, 055104.
- Gamal, R.; Ismail, Y.; Swillam, M.A. Optical biosensor based on a silicon nanowire ridge waveguide for lab on chip applications. J. Opt. 2015, 17, 045802.
- Elsayed, M.Y.; Sherif, S.M.; Aljaber, A.S.; Swillam, M.A. Integrated Lab-on-a-Chip Optical Biosensor Using Ultrathin Silicon Waveguide SOI MMI Device. Sensors 2020, 20, 4955.
- Luan, E.; Shoman, H.; Ratner, D.M.; Cheung, K.C.; Chrostowski, L. Silicon Photonic Biosensors Using Label-Free Detection. Sensors 2018, 18, 3519.
- Zaki, A.O.; Kirah, K.; Swillam, M.A. Integrated optical sensor using hybrid plasmonics for lab on chip applications. J. Opt. 2016, 18, 085803.
- Khalifa, A.E.; Swillam, M.A. Plasmonic silicon solar cells using titanium nitride: A comparative study. JNP 2014, 8, 084098.
- Naik, G.V.; Shalaev, V.M.; Boltasseva, A. Alternative Plasmonic Materials: Beyond Gold and Silver. Adv. Mater. 2013, 25, 3264–3294.
- Naik, G.V.; Kim, J.; Boltasseva, A. Oxides and nitrides as alternative plasmonic materials in the optical range . Opt. Mater. Express OME 2011, 1, 1090–1099.
- Lau, B.; Swillam, M.A.; Helmy, A.S. Hybrid orthogonal junctions: Wideband plasmonic slot-silicon waveguide couplers. Opt. Express OE 2010, 18, 27048–27059.
- Ayoub, A.B.; Ji, D.; Gan, Q.; Swillam, M.A. Silicon plasmonic integrated interferometer sensor for lab on chip applications. Opt. Commun. 2018, 427, 319–325.
- Gamal, R.; Ismail, Y.; Swillam, M.A. Silicon Waveguides at the Mid-Infrared. J. Lightwave Technol. JLT 2015, 33, 3207–3214.
- Elsayed, M.Y.; Ismail, Y.; Swillam, M.A. Semiconductor plasmonic gas sensor using on-chip infrared spectroscopy. Appl. Phys. A 2017, 123, 113.
- Sherif, S.M.; Swillam, M.A. Metal-less silicon plasmonic mid-infrared gas sensor. JNP 2016, 10, 026025.
- Ayoub, A.B.; Swillam, M.A. Silicon Plasmonics On-Chip Mid-IR Gas Sensor. IEEE Photonics Technol. Lett. 2018, 30, 931–934.
- Gamal, R.; Shafaay, S.; Ismail, Y.; Swillam, M.A. Silicon plasmonics at midinfrared using silicon-insulator-silicon platform. JNP 2017, 11, 016006.
- El Shamy, R.S.; Khalil, D.; Swillam, M.A. Mid Infrared Optical Gas Sensor Using Plasmonic Mach-Zehnder Interferometer. Sci. Rep. 2020, 10, 1293.
- Schwarz, B.; Reininger, P.; Ristanić, D.; Detz, H.; Andrews, A.M.; Schrenk, W.; Strasser, G. Monolithically integrated mid-infrared lab-on-a-chip using plasmonics and quantum cascade structures. Nat. Commun. 2014, 5, 4085.
- Moon, G.; Choi, J.-R.; Lee, C.; Oh, Y.; Kim, K.H.; Kim, D. Machine learning-based design of meta-plasmonic biosensors with negative index metamaterials. Biosens. Bioelectron. 2020, 164, 112335.
