Tumor cells evolve in a complex and heterogeneous environment composed of different cell types and an extracellular matrix. Current 2D culture methods are very limited in their ability to mimic the cancer cell environment. Various 3D models of cancer cells have been developed, notably in the form of spheroids/organoids, using scaffold or cancer-on-chip devices. However, these models have the disadvantage of not being able to precisely control the organization of multiple cell types in complex architecture and are sometimes not very reproducible in their production, and this is especially true for spheroids. Three-dimensional bioprinting can produce complex, multi-cellular, and reproducible constructs in which the matrix composition and rigidity can be adapted locally or globally to the tumor model studied. For these reasons, 3D bioprinting seems to be the technique of choice to mimic the tumor microenvironment in vivo as closely as possible.
- 3D bioprinting
- 3D printing
- bioink
- cancer
- cell biology
1. 3D Bioprinting at a Glance
1.1. Introduction
1.2. Bioprinting Technologies
| Type of Technology | Example of Printing Method | Advantages | Disadvantages | Cell Density | Average Cell Viability | Crosslinking | References |
|---|---|---|---|---|---|---|---|
| Droplet-based | Laser | Very high accuracy and resolution Low shear stress Very expensive |
Only low-viscosity bioinks Only 2D patterns (limited high) |
Low (less than 10 million per mL) | High | Depends on biomaterial used | [14][15] |
| Inkjet | High accuracy Low shear stress |
[16][17] | |||||
| Filament-based | Worm drive Pneumatic Syringe/piston |
Large panel of bioinks available Low cost Highly tunable |
Higher shear stress and lower cell viability than other bioprinting technologies | High (more than 10 million per mL) | Medium/high depending on nozzle and pressure | Depends on biomaterial used | [13][18][19][20] |
| Plane-based/Volumetric | DLP/SLA | Fast for large and complex 3D models Very high accuracy |
Few bioinks available Waste of bioink due to its conception |
High (more than 10 million per mL) | High | Photocurable by DLP/SLA technology | [21][22][23][24][25] |
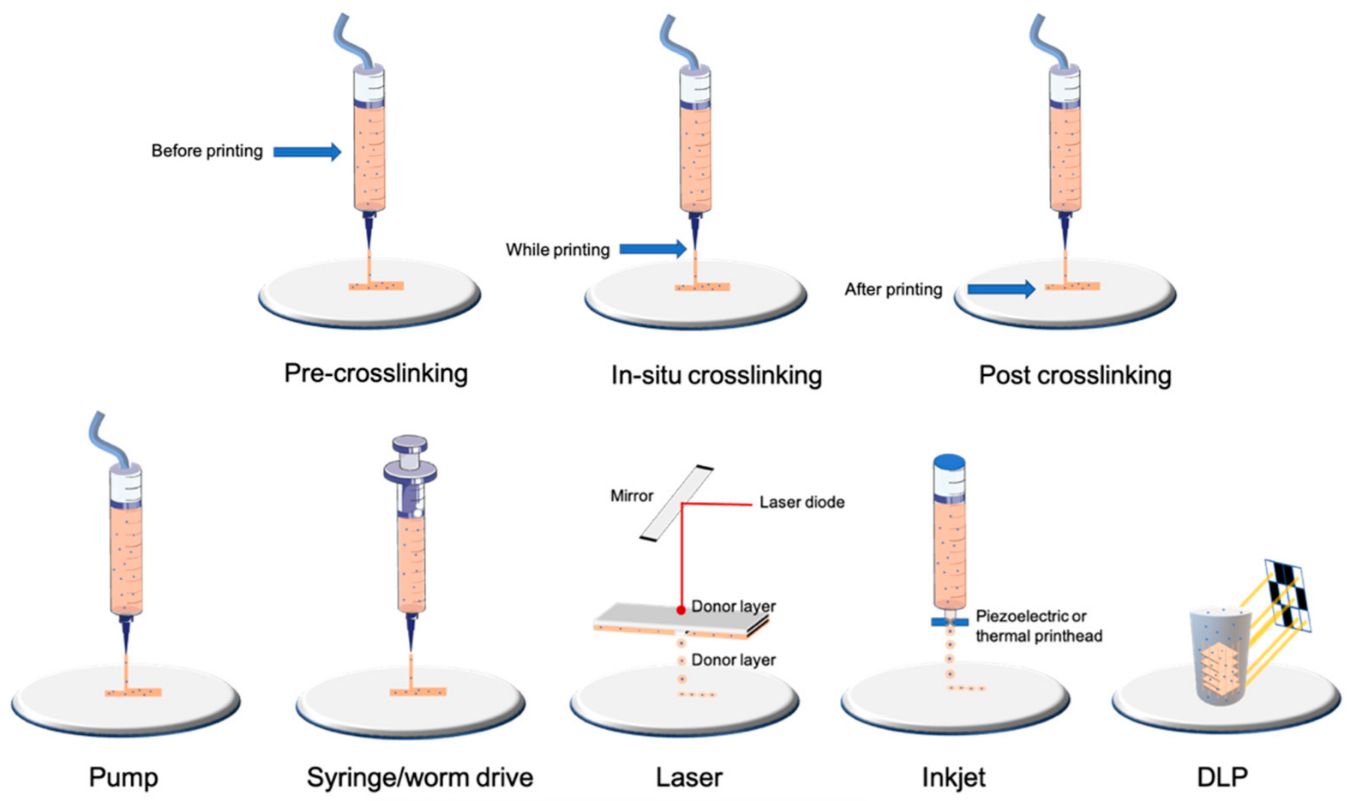
2. Characterization of Cells after Bioprinting
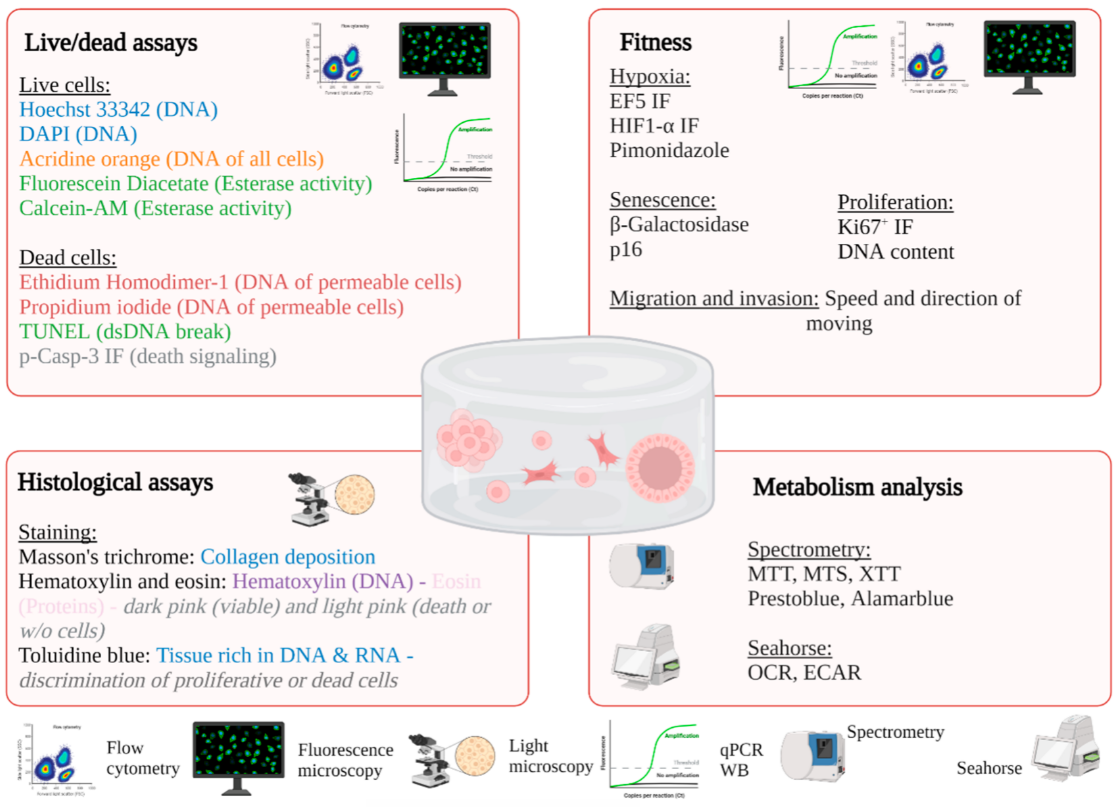
2.1. In Situ Characterization of Cells
2.1.1. Light Microscopy
2.1.2. Fluorescence Microscopy
| Material | Type of Bioink | Bioprinting Technology | Tissue Engineering Model | Cancer Models | Advantages | Drawbacks | Type of Crosslinking | References | |
|---|---|---|---|---|---|---|---|---|---|
| Bioink derived from natural biomaterials | Alginate-based | Natural polysaccharide (brown algae) |
Drop-based Filament-based |
Vascular, cartilage, bone, neural tissue, fibroblast, and many more | Drug delivery Cancer stem cell research Breast cancer, melanoma, and many more cancers Tumor spheroids |
Low cost Good printability Excellent bio-compatibility |
Poor cell adhesion Fast degradation |
Ionic | [46][47][48][49][50][51][52] |
| Gelatin-based | Natural protein (bovine skin and tendon) |
Drop-based Filament-based Plane-based |
Vascular, cartilage, bone, muscle, fibroblast, and many more | Cholangiocarcinoma, bladder cancer, and many more cancers Tumor spheroids |
Excellent bio-compatibility Low-cost High cellular adhesion |
Low viscosity at room or higher temperatures Need a temperature-controlled (cooled printhead) and a cooled printbed Low mechanical strength (higher if blended with methacrylate) |
Chemical Thermal UV Covalent Enzymatic |
[50][53][54][55][56][57][58] | |
| Cellulose and nanocellulose-based | Natural polysaccharide obtained from the biosynthesis of plants or bacteria | Filament-based | Cartilage and bone | Drug delivery Gastric, cervical, pancreatic, and many more cancers |
Great similarity with ECM Excellent bio-compatibility |
Low viscosity for cellulose nanocrystals Mainly used mixed with other natural biomaterials |
Enzymatic UV |
[59][60][61][62][63] | |
| Matrigel | Solubilized basement membrane matrix secreted by Engelbreth-Holm-Swarm (EHS) mouse sarcoma cells | Filament-based Drop-based |
Vascular, liver, bone, lung, and many more | Tumor spheroids Many types of cancer |
Most used material in cancer research Excellent bio-compatibility Very well characterized for organoid/spheroid formation |
Cannot be used alone due to its complex rheological behavior and low mechanical properties Limited use in vivo due to its mouse tumor origin Expensive High batch variability |
Thermal | [64][65][66][67][68] | |
| Collagen-I-based | Natural protein (rat tail or bovine skin and tendon) | Drop-based Filament-based |
Hard tissues (bone, osteochondral, cartilage) Skin, cardiovascular, and liver tissues; nervous system; and cornea |
Tumor spheroids Neuroblastoma, breast cancer |
Excellent bio-compatibility High cellular adhesion Minimal immunogenicity Excellent printability Enzymatically degradableMechanical and structural properties close to native tissue |
Low shape fidelity | pH Thermal |
[69][70][71][72] | |
| Hyaluronic-acid-based | Natural polysaccharide (bacterial fermentation or animal products) | Filament-based | Hard tissues (bone, osteochondral, cartilage) | Tumor spheroids Melanoma, breast cancer |
Excellent bio-compatibility Highly tunable (wide variety and high degree of potential chemical modifications) Interact with cell receptors |
Poor mechanical strength Mainly used mixed with other natural biomaterials |
Depends on the other biomaterial/chemical modifications Physical or covalent |
[73][74][75][76] | |
| Agarose-based | Natural polysaccharide derived from red seaweed | Filament-based | Bone, vascular, neural, and adipose tissue | Leukemia | Good biocompatibility Great similarity with ECM Thermo-reversible gelling |
Poor cell survival if not blended with another biomaterial Poor printability (needs high temperature for dispensing (70 °C) and gels at low temperatures) |
Thermal Ionic |
[65][77][78] | |
| Fibrin-based | Natural protein (human plasma) |
Filament-based Drop-based |
Muscular, neural, skin, and adipose tissue, wound healing model | Drug release Glioblastoma |
High shape fidelity (depending on fibrinogen–thrombin concentration) Excellent biocompatibility Enzymatically degradable |
Medium cell adhesion Low mechanical properties |
Enzymatic (fibrinogen–thrombin) | [79][80][81] | |
| Silk-derived | Natural protein (bombyx mory) |
Filament-based | Hard tissues (bone, osteochondral, cartilage), vascular tissue | Drug delivery | High shape fidelity Low Cost Good biocompatibility |
Lacks cell-binding domains Medium cell viability Needs other supportive material for cell proliferation (alginate, gelatin, etc.) Poor printability performance |
Enzymatic Physical |
[82][83][84][85] | |
| Gellan gum | Natural polysaccharide | Filament-based | Hard tissues (bone, osteochondral, cartilage), brain-like structures | Drug delivery | Excellent biocompatibility Low cost Rapid gelation |
Poor printability performance | Thermal | [86][87][88] | |
| Chitosan | Natural polysaccharide produced by deacetylation of chitin (extract from shrimps) | Filament-based Drop-based Plane-based |
Hard tissues (bone, osteochondral, cartilage), vascular, skin, and hepatic tissues | Drug delivery | Good biocompatibility Medium to high cell viability |
Medium cell adhesion Low shape fidelity Low mechanical properties |
Ionic UV |
[89][90][91] | |
| Polypeptides | Corning (PuraMatrix) | Filament-based Droplet-based Plane-based |
Liver, neural | Ovarian cancer | Self-assembly Adapted for soft-tissue applications and in conjunction with other materials |
Low pH leading to low cell viability | Ionic-complementary self-assembly | [92][93] | |
| De-cellularized matrix-based (dECM) | Natural matrix | Filament-based | Adipose, hepatic, and heart tissues; MSCs; cancer models | Many tumor models depending on dECM | Renders natural ECM Tissue-specific |
Low mechanical properties Protein denaturation during fabrication processes Poor printability if not mixed with another biomaterial Long procedure |
Depends on the other biomaterial/chemical modifications | [94][95][96][97] | |
| Bioink derived from synthetic biomaterials | AM (acrylamide) | Polyacrylamide | Filament-based Plane-based Droplet-based |
Different stiffness models | Melanoma, breast cancer | Wide range of elasticity Most standardized protocol |
Suitable for 2D culture only or necessary to couple it with another material | UV | [98][99] |
| PCL/PLGA | Poly(caprolactone)/Poly(lactic–glycolic acid) | Filament-based Drop-based |
Hard tissues (bone, osteochondral, cartilage) | Mainly depends on the natural biomaterial used | Good mechanical strength Controllable rate of degradation |
Mainly used as a scaffold (melting temperature around 60 °C not compatible with cell viability) Needs other supportive material for cell proliferation (alginate, gelatin, etc.) |
Depends on the natural biomaterial used | [100][101][102] | |
| PEG | Polymer of ethylene oxide | Filament-based | Vascular and bone tissue | Highly tunable (mechanical properties, polymerization, chemical composition) | Needs chemical modification to be printed Requires the addition of bioactive molecules to allow cellular interaction (high hydrophobicity) |
UV if mixed with a photoinitiator Condensation Michael-type addition Click chemistry Native chemical ligation Enzymatic reaction |
[103][104][105] | ||
| Pluronic | Triblock copolymer of poly(ethylene glycol)-poly(propylene oxide)-poly(propylene glycol) | Filament-based | Cartilage | High shape fidelity Good printability |
Lacks cell-binding domains Low cell viability Poor mechanical strength |
Covalent | [106][107] | ||
| PU | Polyurethane | Filament-based | Cartilage Neural stem cells |
Good biocompatibility and biodegradability High mechanical strength |
Needs other supportive material for cell proliferation (alginate, gelatin, etc.) | Depends on the natural biomaterial used | [108][109] |
2.1.3. Electronic Microscopy
2.2. Characterization of Cells after Isolation or Lysis
2.2.1. Molecular Biology
2.2.2. Flow Cytometry
| Methods | Description | Pros and Cons | Markers | REF | |
|---|---|---|---|---|---|
| Microscopy | |||||
| Light | Phase contrast | Monitoring of proliferation and morphology of cells | +: • Nondestructive • No markers are added • Low cost • Easy with transparent gels (GelMA, matrigel) −: • No possibility to identify subcellular structures • Difficult with opaque or non-transparent gels (e.g.,: alginate with nanocellulose) |
Not suitable | [31][32][33] |
| Bright field | The transmission of light is more or less attenuated depending on the density or marking of the sample | +: • Suitable for large samples −: • Requires histological staining • Preparation of sample • Quantification of thick sample |
Hematoxylin–eosin Masson’s trichrome Trypan blue |
[32][33][34] | |
| Fluorescence | LSM Epifluorescence Confocal |
The use of a fluorescent marker is necessary to highlight a subcellular structure; possibility of monitoring structures over time (if vital markers) | +: • Monitoring of many possible structures −: • Requires cutting for oversized constructions for epi and confocal microscopy • Need to fix for certain markers • Important autofluorescence for chitosan or alginate/cellulose hydrogels in UV |
Live/dead staining Or calcein AM/propidium iodide Or ethidium homodimer Active-caspase3/7 green Hoechst 33342 HIF1-α, Ki67 |
[39][40][41][42][123] |
| Electronic | Scanning | Surface is scanned with a beam of electrons, emitted signal provides images | +: • High resolution −: • The preparation procedure is tedious • Frequent preparation artifacts (collapse) |
Not suitable | [33] |
| Transmission | The part of beam of electrons is transmitted into specimens allowed to obtain images | Not suitable | [33][110] | ||
| Flow cytometry | |||||
| Flow cytometry | Analysis of physical parameters (size and granularity) for each cell but also the level of fluorescence | +: • Quantitative analysis −: • Disaggregation can be a problem • Necessity to have a large cell number due to loss of cells during dissociation |
7-AAD CFSE |
[33][117] | |
| Spectroscopy | |||||
| Spectrometry or fluorimetry | Production or utilization of a fluorescent or chromatic compound | +: • Well-described for 2D culture and frequently used • Can be used for kinetic monitoring −: • Ensure that the efficiency is adapted for 3D |
ACP, LDH, prestoblue, alamar blue, DNA content | [43][124][125][126] | |
| Molecular biology | |||||
| RTqPCR Western blot |
Quantification of gene expression at mRNA or protein level | +: • Quantitative analysis • Easier by using the enzymatic method on natural inks (e.g., collagenase for GelMA or ColMA, hyaluronidase for hyaluronic acid) −: •Adaptation of the homogenization and extraction protocol to obtain an adequate quantity and quality of RNA/proteins for analyses |
Bax/Bcl2 HIF1-α, Ki67 |
[34][110][119] | |
| Metabolism | |||||
| GC–MS (Gas chromatography–mass spectrometry) | Detection of molecules of interest according to their mass/charge ratio after ionization | +: • Considerably less cellular material compared to NMR, high sensitivity, −: • Use of radioisotopes, complex sample preparation, high cost |
13C-Glucose | [127][128] | |
| NMR (nuclear magnetic resonance) spectroscopy | Determination of the composition of a sample by applying a magnetic field via the orientation of the nuclear spins of the atoms | +: • High reproducibility, sample can be analyzed directly, low cost −: • Use of radioisotopes, low sensitivity |
[129][130] | ||
| PET scan (positron emission tomography) | Injection of a radiographic tracer and monitoring by imaging to detect localization of [18F]FDG | +: • Classically used in medicine, monitoring over time −: • Low resolution (1.5 mm) |
[18F]FDG | [125][131] | |
| Seahorse | Quantification of the oxygen consumption rate (OCR) and the extracellular acidification rate (ECAR) | +: • High sensitivity (from 5000 cells, theoretically), possibility to test many conditions in parallel −: • Difficulties in normalizing results, limited number of injections, limited sample thickness |
Not suitable | [132][133] | |
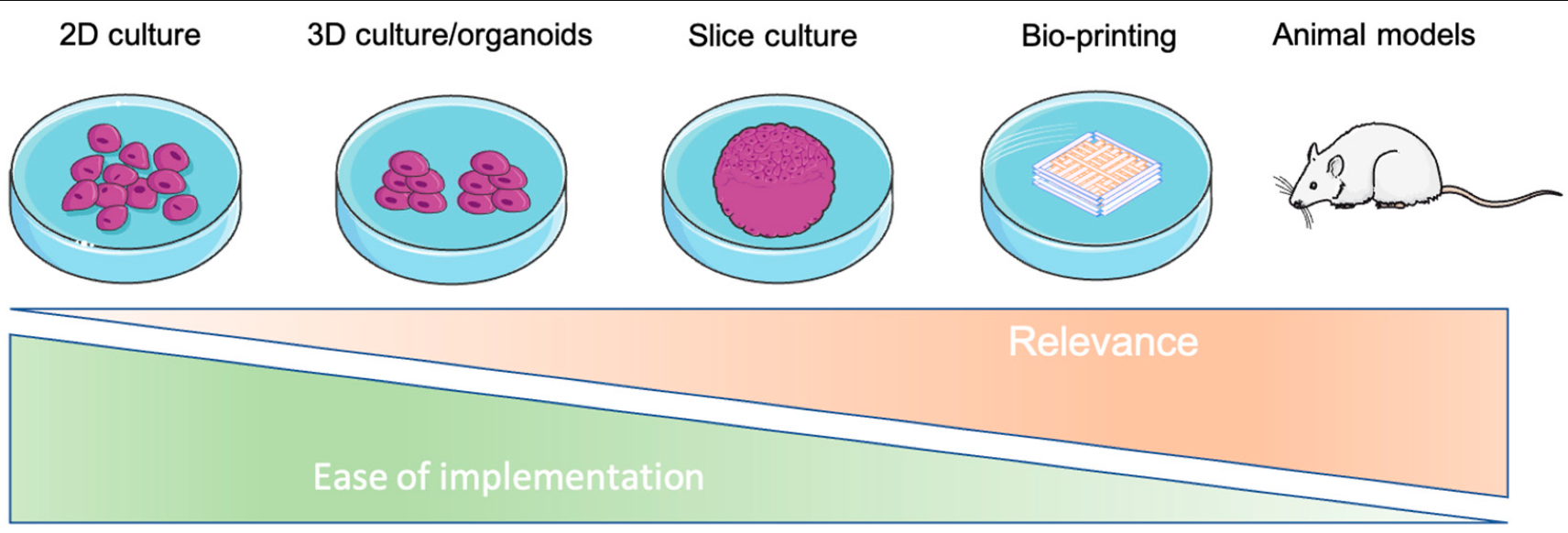
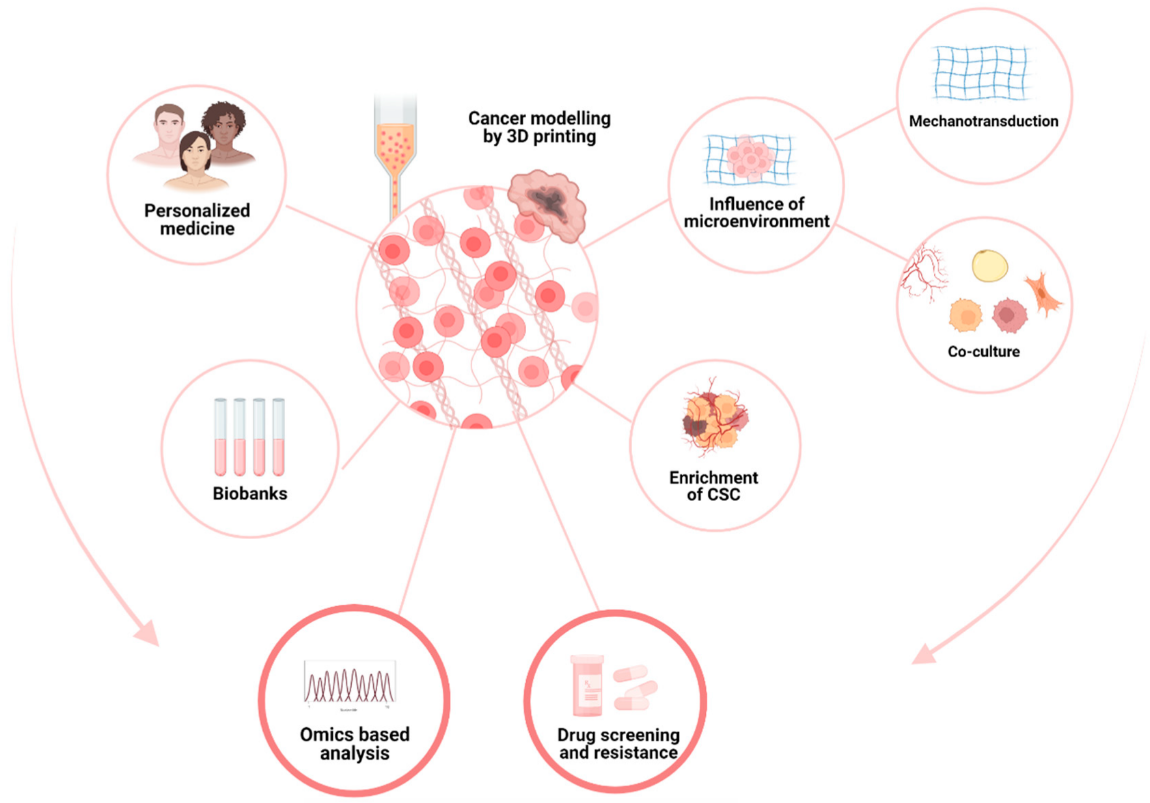
2.3. Recapitulate Cancer’s Relation to the Microenvironment
2.3.1. Cells–ECM Interaction
2.3.2. Neoangiogenesis
2.4. Mechanical Environment
2.4.1. Mechanotransduction
This entry is adapted from the peer-reviewed paper 10.3390/ijms23073432
