Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Biology
Mitochondria play a critical role in providing energy, maintaining cellular metabolism, and regulating cell survival and death. To carry out these crucial functions, mitochondria employ more than 1500 proteins, distributed between two membranes and two aqueous compartments. An extensive network of dedicated proteins is engaged in importing and sorting these nuclear-encoded proteins into their designated mitochondrial compartments. Defects in this fundamental system are related to a variety of pathologies, particularly engaging the most energy-demanding tissues.
- mitochondria
- mitochondrial protein import
1. Introduction
In general, mitochondria are known as the powerhouses of cells since their major function is to produce ATP as an energy source. Besides that, mitochondria are required for the regulation of calcium homeostasis, lipid and amino acid metabolism, and biosynthesis of heme and iron-sulfur complexes. Over the last years, it has become evident that mitochondria are signaling organelles, which regulate processes like apoptotic cell death as well as nuclear transcription [1].
Mitochondrial functions decline with the aging process of the organism, as mitochondria are subjected to a variety of biochemical stress conditions. Those lead to the accumulation of damaged molecules that directly or indirectly interfere with the regular biochemical processes occurring in mitochondria. Most prominent in this context are oxidative stress reactions caused by the accumulation of reactive oxygen species (ROS). ROS are generated as byproducts during the metabolic reactions of the OXPHOS process, in addition to detrimental environmental impacts [2]. Mitochondrial biogenesis capacity declines at older age, in particular due to the accumulation of mutations in the mitochondrial and nuclear genome. These mutations lead to the generation of defective or aberrant enzymatic components of the mitochondrial structure and metabolism, resulting in mitochondrial and eventually cellular dysfunction. This general problem is exacerbated by a relatively error-prone mitochondrial genome replication and inefficient repair processes [3].
As cellular survival depends on the maintenance of protein function, also called proteostasis, an efficient protein biogenesis process is a prerequisite to providing the required proteins for all cellular functions. Due to the dual genetic origin of mitochondrial proteins, a portion is encoded in the mitochondria, but most are nuclear-encoded; efficient protein biosynthesis and subsequent import into the organelle are essential for mitochondrial function. In addition to the transport processes into the organelle, mitochondrial polypeptides—irrespective of their source—need to undergo proper folding and assembly into active enzymes or enzyme complexes. Thus, the biogenesis of mitochondrial proteins represents a very complex process that is also prone to errors and defects correlating with diseases. Of note, not only the proteins in the inner (IMM) and outer mitochondrial membranes (OMM) are required for proper complex assembly, but also the phospholipid composition of the mitochondrial membranes plays a critical role. For example, it has been demonstrated that the lipid composition changes with age; in particular, the content of the mitochondria-specific lipid cardiolipin declines, which may play a central role in age-related diseases [4]. Age-related accumulation of cholesterol in mitochondria is a proposed trigger of Alzheimer’s disease (AD) [5,6].
Stress- or age-related damage of individual proteins or even the full organelle is counteracted by an array of different protective processes. On the protein side, a variety of chaperone and protease enzymes help to refold or remove damaged polypeptides before they accumulate and induce malfunction. To deal with unfolded polypeptides, mitochondria contain the full set of Hsp70- and Hsp60-type chaperones, similar to their bacterial ancestors [7]. In addition, each mitochondrial subcompartment, including the membranes, contains protease enzymes that specifically digest polypeptides that could not be refolded or assembled into their respective enzyme complexes [8]. Failure of this protein quality control (PQC) system contributes to many human diseases [9]. Specific signaling processes from mitochondria to the nucleus, summarized under the expression “unfolded protein response (mtUPR)”, increase the mitochondrial PQC capacity by enhancing the expression of the respective chaperones and proteases. If these protective processes fail, defective mitochondria themselves seem to be removed by a regulated and specific form of autophagy, named mitophagy [10].
As mitochondrial functions and quality control are largely dependent on the import of proteins produced in the cytoplasm, disruptions in import create cascading effects in mitochondria leading to diminished metabolic function, increased production of ROS, and failures in regulated cell death response [11]. Mitochondrial dysfunction is increased particularly with older age. For several diseases, most importantly affecting organs that particularly depend on mitochondria, namely the brain and the heart, mitochondrial dysfunction has been broadly described [1,12,13,14,15].
Not surprisingly, a number of recent reviews describe different aspects of mitochondrial function and structure, in particular mitochondrial machineries for protein import and assembly [16]. Considerably fewer reviews can be found on the connection between mitochondrial protein import and aging-related diseases [17].
2. Mitochondrial Protein Import Machinery
Nuclear-encoded mitochondrial proteins typically contain signaling sequences that are recognized in the cytosol by the mitochondrial protein import machinery and transported into mitochondria in an ATP-dependent manner (for review, see [16]). Mitochondrial targeting sequences can either be N-terminal (e.g., subunit 8 of cytochrome c oxidase), internal (e.g., ADP/ATP carrier), or C-terminal (e.g., VDAC) [18,19,20,21]). N-terminal presequences are the most frequent and can be recognized by mitochondria import machinery either during or after translation [22]. After import, around 70% of presequences are cleaved by mitochondrial processing peptidases (MPP) [23]. Internal and C-terminal mitochondrial targeting sequences can only be recognized after protein translation is complete. Mitochondrial proteins employing internal or C-terminal targeting sequences are frequently hydrophobic and require assistance from chaperones such as Hsp70/90 to prevent misfolding and aggregation in the cytoplasm.
Mitochondrial protein recognition occurs at the translocase of the outer membrane (TOM) complex. The TOM component TOMM20 acts as a direct receptor for N-terminal signal sequences, while TOMM70 seems to be dedicated to the recognition of hydrophobic proteins with internal signal sequences. These hydrophobic proteins reach the mitochondrial surface, often bound by cytosolic chaperones that also can interact with TOMM70 [16]. After passing through pore formed by the TOM complex, proteins are sorted between TIM22, TIM23, SAM, or MIA machinery, depending on the protein destination and fold (Figure 1). Insertase OXA1L inserts mitochondria encoded proteins into IMM. In yeast, mitochondrial insertase Oxa1p additionally assists in inserting multi α-helical nuclear-encoded proteins, but the same behavior for human OXA1L was not yet verified [24,25].
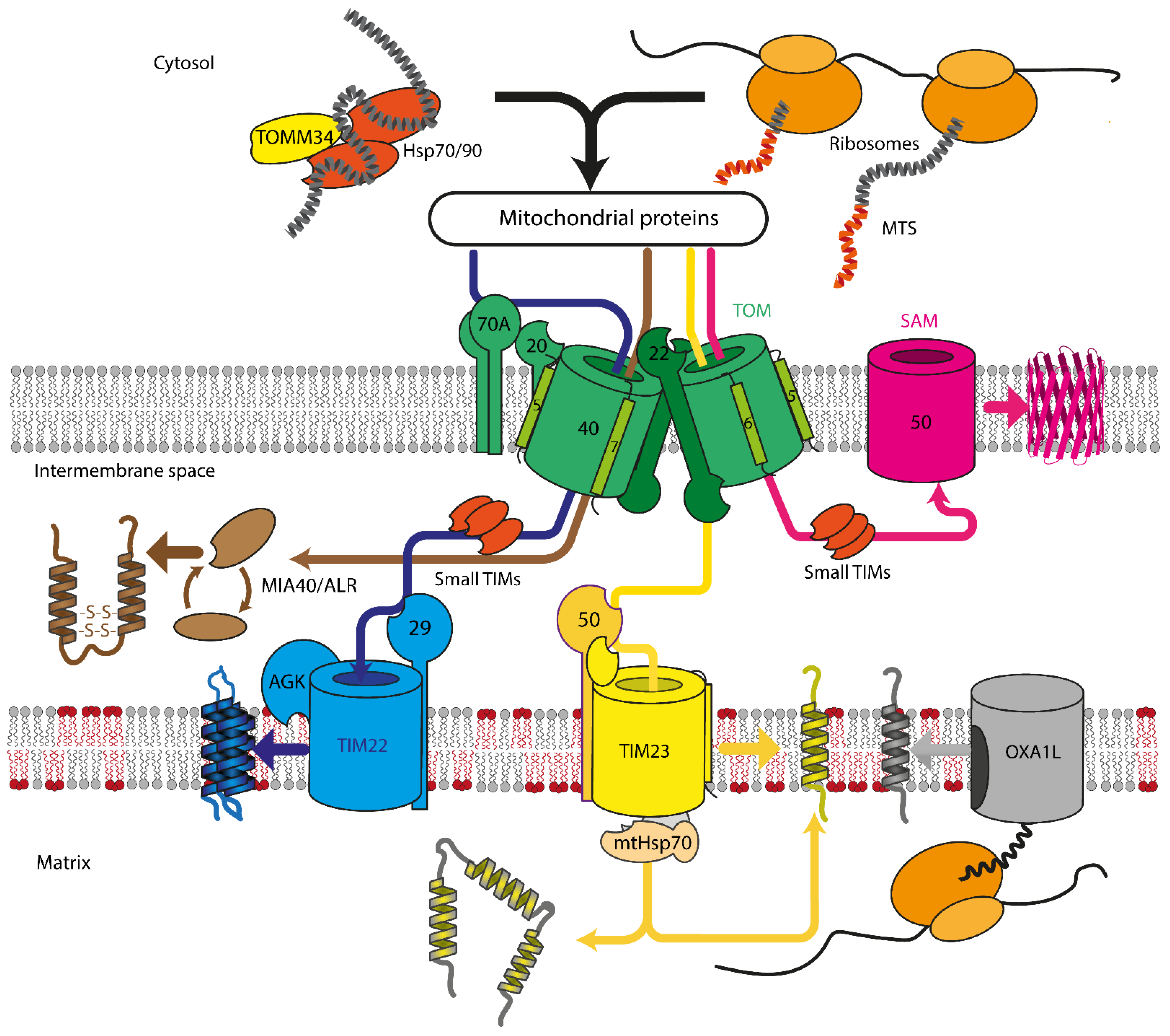
Figure 1. Overview of mitochondrial protein import in mammalian cells. Proteins are recognized by TOMM70A/TOMM20/TOMM22 and are imported either co- or post-translationally through the TOM complex, containing TOMM40, TOMM22, TOMM5, TOMM6, and TOMM7. Hydrophobic proteins employ Hsp70/90 complex with participation of TOMM34 to prevent misfolding in the cytoplasm. Inside the intermembrane space, depending on the nature and destination of the precursor protein, proteins are delivered to different compartments. β-barrels of the outer membrane are inserted into the outer mitochondrial membrane by SAM complex. Intermembrane space proteins with cysteine motifs are oxidized to the mature form by the MIA40/ALR system. Metabolite carriers are inserted into IMM by TIM22 complex, composed of TIMM22, TIMM29, and acylglycerol kinase (AGK). Other IMM and matrix proteins are inserted/transported by TIM23 complex. Primary TIMM23 pore is associated with TIMM50 (recognizes signals and interacts with TOM complex), TIMM44 with associated mtHsp70 (forming presequence-associated motor helping matrix protein transfer), or TIMM21 for protein release into IMM. Mitochondrial-encoded proteins are inserted into the IMM by OXA1L insertase from the matrix side.
In addition to signaling sequences, mitochondrial protein import efficiency is further enhanced by localization of mRNA encoding mitochondrial proteins near the OMM and its translation in the vicinity of the translocation machinery [26,27,28], thus bypassing the need for intermediate chaperone-mediated transport. The observed mRNA localization near OMM is guided either by the interaction of protein nascent chains with TOM complex or by the interaction between 3′ mRNA end, specific RNA-binding proteins (RBPs), and OMM proteins. In human cells, RBPs are only incompletely described, but some proteins were found to co-localize with mRNA on the OMM surface, e.g., CluH and PUM [29,30]. Recent advances in proximity labeling identified several new RBPs [31] on the OMM surface, including SYNJ2BP, which persists on the OMM surface after disruption of protein synthesis and binds several mRNAs of OXPHOS proteins. RBPs’ participation in mitochondrial protein transport presents another potential avenue of protein import disruption caused by mutations/aging/environmental factors.
This entry is adapted from the peer-reviewed paper 10.3390/cells10123528
This entry is offline, you can click here to edit this entry!
