The eukaryotic translation initiation factor 5A (eIF5A) is an evolutionarily conserved protein that binds ribosomes to facilitate the translation of peptide motifs with consecutive prolines or combinations of prolines with glycine and charged amino acids. It has also been linked to other molecular functions and cellular processes, such as nuclear mRNA export and mRNA decay, proliferation, differentiation, autophagy, and apoptosis. The growing interest in eIF5A relates to its association with the pathogenesis of several diseases, including cancer, viral infection, and diabetes. It has also been proposed as an anti-aging factor: its levels decay in aged cells, whereas increasing levels of active eIF5A result in the rejuvenation of the immune and vascular systems and improved brain cognition. Data have linked the role of eIF5A in some pathologies with its function in maintaining healthy mitochondria. The eukaryotic translation initiation factor 5A is upregulated under respiratory metabolism and its deficiency reduces oxygen consumption, ATP production, and the levels of several mitochondrial metabolic enzymes, as well as altering mitochondria dynamics. However, although all the accumulated data strongly link eIF5A to mitochondrial function, the precise molecular role and mechanisms involved are still unknown.
- eIF5A
- mitochondria
- translation
- spermidine
- OXPHOS
- TCA
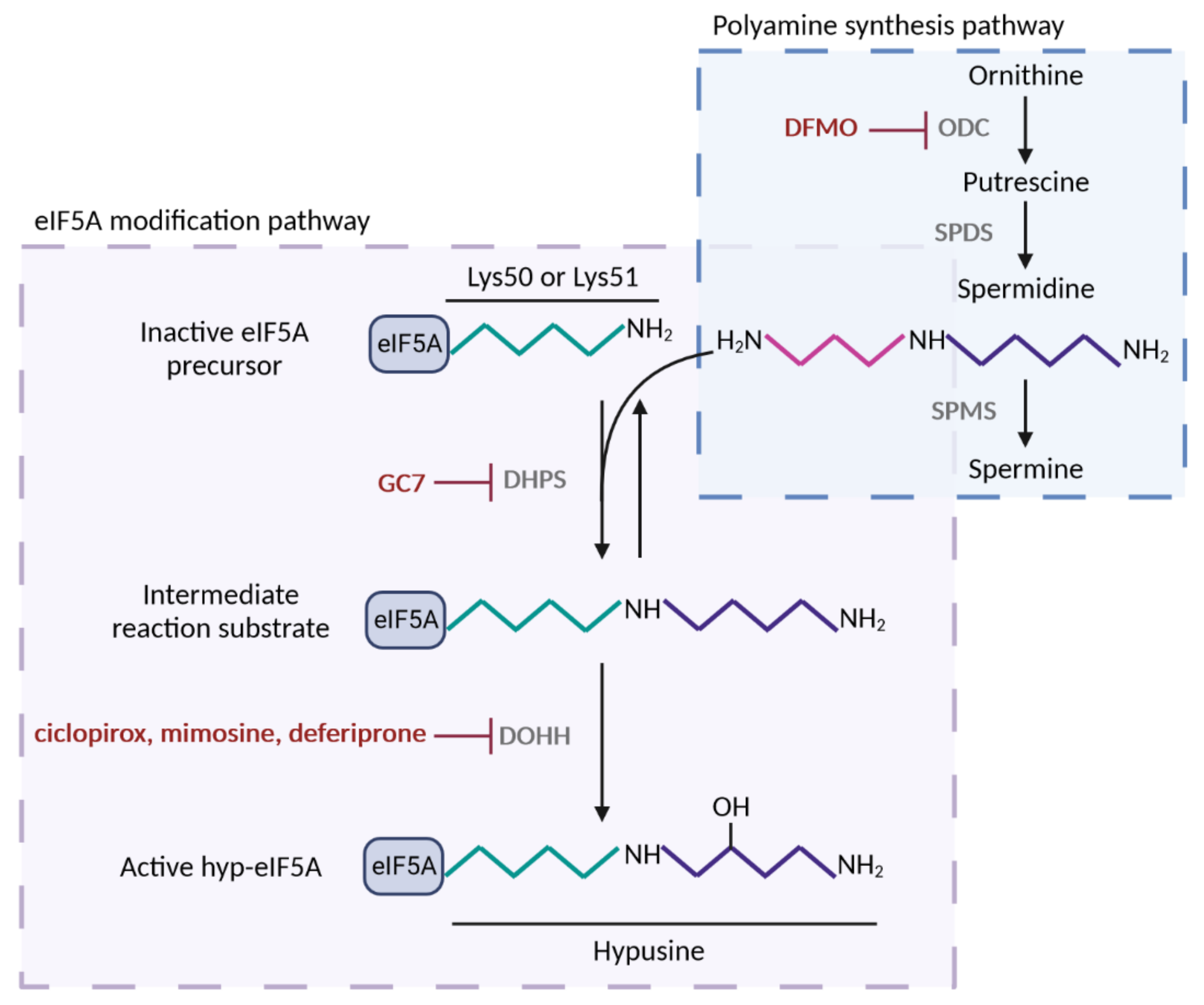
1. The Molecular Function of eIF5A
2. Identification of Mitochondrial Processes and Targets under the Control of eIF5A
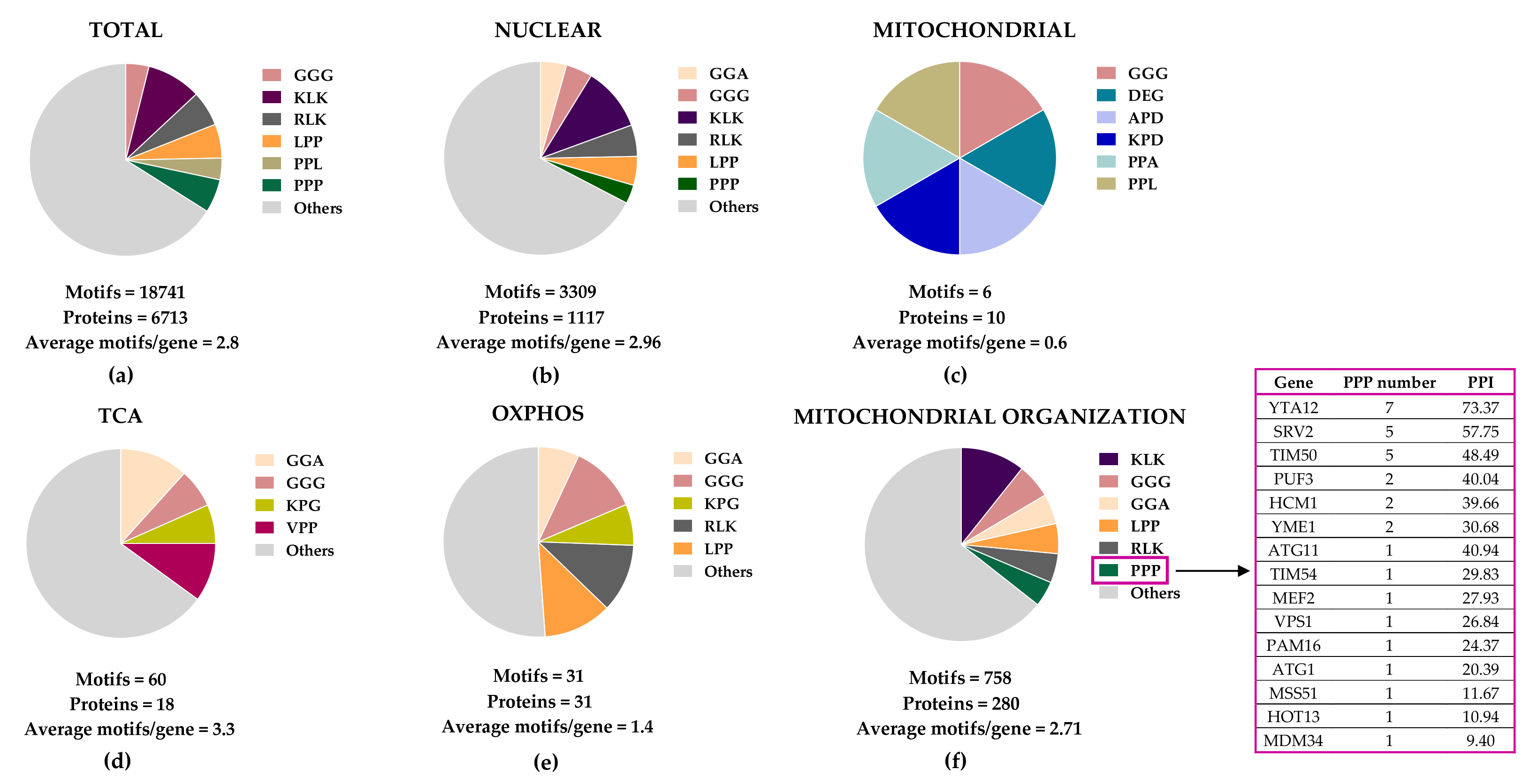 Figure 2. Distribution of eIF5A-dependent motifs in Saccharomyces cerevisiae mitochondrial proteins. Distribution of the 43 highest-scoring eIF5A-dependent ribosome-pausing tri-peptide motifs [11] in the proteins of the whole yeast genome (a), nuclear-encoded mitochondrial proteins (b), mitochondrial-encoded proteins (c), the tricarboxylic acid (TCA) cycle (d), oxidative phosphorylation (OXPHOS) (e), and in the mitochondrial organization Gene Ontology functional category (f). The table shows the proteins involved in mitochondrial organization with at least one PPP motif. The protein pause index (PPI) is calculated as the sum of the quantitative value of the ribosome pause provoked by depletion of eIF5A in each of the 43 highest eIF5A-dependent tri-peptide motifs [11] found in the amino acid sequence of each protein, and is higher in proteins that are putatively more dependent on eIF5A for their translation.
Figure 2. Distribution of eIF5A-dependent motifs in Saccharomyces cerevisiae mitochondrial proteins. Distribution of the 43 highest-scoring eIF5A-dependent ribosome-pausing tri-peptide motifs [11] in the proteins of the whole yeast genome (a), nuclear-encoded mitochondrial proteins (b), mitochondrial-encoded proteins (c), the tricarboxylic acid (TCA) cycle (d), oxidative phosphorylation (OXPHOS) (e), and in the mitochondrial organization Gene Ontology functional category (f). The table shows the proteins involved in mitochondrial organization with at least one PPP motif. The protein pause index (PPI) is calculated as the sum of the quantitative value of the ribosome pause provoked by depletion of eIF5A in each of the 43 highest eIF5A-dependent tri-peptide motifs [11] found in the amino acid sequence of each protein, and is higher in proteins that are putatively more dependent on eIF5A for their translation.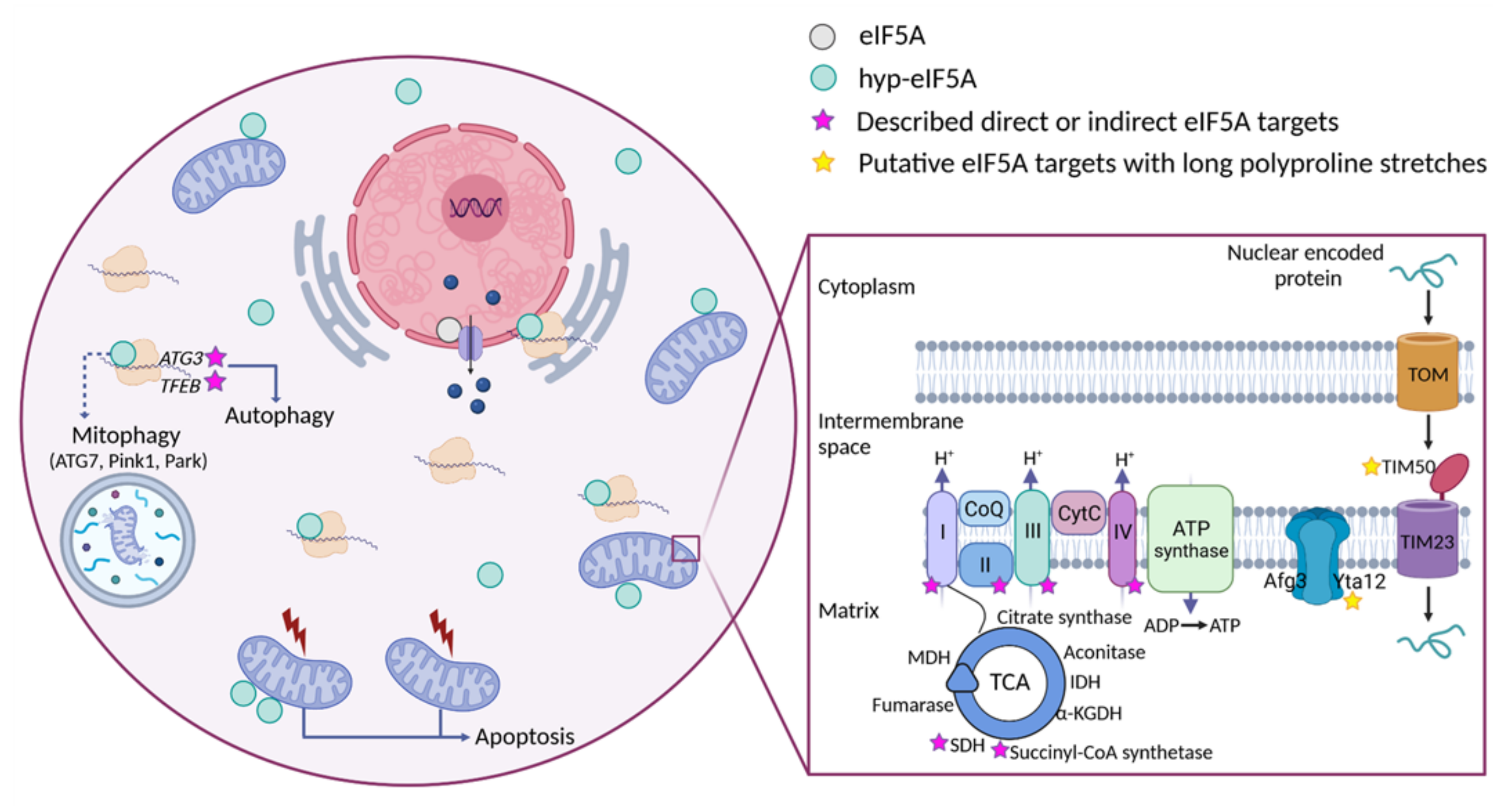 Figure 3. Cellular funtions of eIF5A and model for its role in maintaining mitochondrial activity. eIF5A is known to be implicated in different cellular processes, although the most relevant and mitochondrial-related of these are represented in the Figure. Bound to ribosomes, hyp-eIF5A facilitates translation elongation at specific motifs [10][11], as well as ER-coupled translation [23][25][26]. In the nucleus, eIF5A helps to export certain mRNAs and proteins [76]. eIF5A plays a controversial role in apoptosis, as it has been defined to be necessary to induce the mitochondrial mediated apoptosis [77][78], but also to lead to cell death when inhibited [79]. Hyp-eIF5A promotes autophagy through the translation of the autophagy factors ATG3 (autophagy-related 3) and TFEB (transcription factor EB) [42][43]. Increasing evidence shows a direct link between hyp-eIF5A and mitochondrial function. In addition to its association with mitochondria [80][81][82][78], some proteins of both the TCA and ETC have been described to be directly or indirectly affected under hyp-eIF5A inhibition [83][84]. It has also been proposed that hyp-eIF5A could mediate mitophagy through ATG7 (Autophagy Related 7), Pink1 (the mitophagy-associated PTEN-induced putative kinase), and Park (the E3 ubiquitin ligase Parkin) proteins [85]. Other proteins involved in the mitochondrial transport of nuclear-encoded proteins and mitochondrial organization are considered as putative eIF5A targets (Figure 2). Among these, the mitochondrial inner-membrane integral proteins Yta12 (protease of the Yta12/Afg3 complex and yeast homolog of human AFG3L2), and Tim50 (essential subunit of the TIM23 complex and yeast homolog of human TIMM50) contain long polyproline stretches in their amino acids sequences, suggesting a possible dependence on eIF5A for their translation and, thus, a possible link between hyp-eIF5A and mitochondrial function. Figure processing was carried out using BioRender software.
Figure 3. Cellular funtions of eIF5A and model for its role in maintaining mitochondrial activity. eIF5A is known to be implicated in different cellular processes, although the most relevant and mitochondrial-related of these are represented in the Figure. Bound to ribosomes, hyp-eIF5A facilitates translation elongation at specific motifs [10][11], as well as ER-coupled translation [23][25][26]. In the nucleus, eIF5A helps to export certain mRNAs and proteins [76]. eIF5A plays a controversial role in apoptosis, as it has been defined to be necessary to induce the mitochondrial mediated apoptosis [77][78], but also to lead to cell death when inhibited [79]. Hyp-eIF5A promotes autophagy through the translation of the autophagy factors ATG3 (autophagy-related 3) and TFEB (transcription factor EB) [42][43]. Increasing evidence shows a direct link between hyp-eIF5A and mitochondrial function. In addition to its association with mitochondria [80][81][82][78], some proteins of both the TCA and ETC have been described to be directly or indirectly affected under hyp-eIF5A inhibition [83][84]. It has also been proposed that hyp-eIF5A could mediate mitophagy through ATG7 (Autophagy Related 7), Pink1 (the mitophagy-associated PTEN-induced putative kinase), and Park (the E3 ubiquitin ligase Parkin) proteins [85]. Other proteins involved in the mitochondrial transport of nuclear-encoded proteins and mitochondrial organization are considered as putative eIF5A targets (Figure 2). Among these, the mitochondrial inner-membrane integral proteins Yta12 (protease of the Yta12/Afg3 complex and yeast homolog of human AFG3L2), and Tim50 (essential subunit of the TIM23 complex and yeast homolog of human TIMM50) contain long polyproline stretches in their amino acids sequences, suggesting a possible dependence on eIF5A for their translation and, thus, a possible link between hyp-eIF5A and mitochondrial function. Figure processing was carried out using BioRender software.This entry is adapted from the peer-reviewed paper 10.3390/ijms23031284
References
- Schnier, J.; Schwelberger, H.G.; Smit-McBride, Z.; Kang, H.A.; Hershey, J.W. Translation initiation factor 5A and its hypusine modification are essential for cell viability in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 1991, 11, 3105–3114.
- Kulak, N.A.; Pichler, G.; Paron, I.; Nagaraj, N.; Mann, M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods 2014, 11, 319–324.
- Kemper, W.M.; Berry, K.W.; Merrick, W.C. Purification and properties of rabbit reticulocyte protein synthesis initiation factors M2Bα and M2Bβ. J. Biol. Chem. 1976, 251, 5551–5557.
- Benne, R.; Hershey, J.W.B. The mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. J. Biol. Chem. 1978, 253, 3078–3087.
- Jao, D.L.E.; Chen, K.Y. Tandem affinity purification revealed the hypusine-dependent binding of eukaryotic initiation factor 5A to the translating 80S ribosomal complex. J. Cell. Biochem. 2006, 97, 583–598.
- Zanelli, C.F.; Maragno, A.L.C.; Gregio, A.P.B.; Komili, S.; Pandolfi, J.R.; Mestriner, C.A.; Lustri, W.R.; Valentini, S.R. eIF5A binds to translational machinery components and affects translation in yeast. Biochem. Biophys. Res. Commun. 2006, 348, 1358–1366.
- Gregio, A.P.B.; Cano, V.P.S.; Avaca, J.S.; Valentini, S.R.; Zanelli, C.F. eIF5A has a function in the elongation step of translation in yeast. Biochem. Biophys. Res. Commun. 2009, 308, 785–790.
- Saini, P.; Eyler, D.E.; Green, R.; Dever, T.E. Hypusine-containing protein eIF5A promotes translation elongation. Nature 2009, 459, 118–121.
- Gutierrez, E.; Shin, B.S.; Woolstenhulme, C.J.; Kim, J.R.; Saini, P.; Buskirk, A.R.; Dever, T.E. eif5A promotes translation of polyproline motifs. Mol. Cell 2013, 51, 35–45.
- Schuller, A.P.; Wu, C.C.C.; Dever, T.E.; Buskirk, A.R.; Green, R. eIF5A Functions Globally in Translation Elongation and Termination. Mol. Cell 2017, 66, 194–205.
- Pelechano, V.; Alepuz, P. EIF5A facilitates translation termination globally and promotes the elongation of many non polyproline-specific tripeptide sequences. Nucleic Acids Res. 2017, 45, 7326–7338.
- Park, M.H.; Wolff, E.C. Hypusine, a polyamine-derived amino acid critical for eukaryotic translation. J. Biol. Chem. 2018, 293, 18710–18718.
- Park, M.H.; Lee, Y.B.; Joe, Y.A. Hypusine is essential for eukaryotic cell proliferation. NeuroSignals 1997, 6, 115–123.
- Klier, H.; Csonga, R.; João, H.C.; Eckerskom, C.; Auer, M.; Lottspeich, F.; Eder, J. Isolation and Structural Characterization of Different Isoforms of the Hypusine-Containing Protein eIF-5A from HeLa Cells. Biochemistry 1995, 34, 14693–14702.
- Schwelberger, H.G.; Kang, H.A.; Hershey, J.W.B. Translation initiation factor eIF-5A expressed from either of two yeast genes or from human cDNA. Functional identity under aerobic and anaerobic conditions. J. Biol. Chem. 1993, 268, 14018–14025.
- Zarembinski, T.I.; Hung, L.I.W.; Mueller-Dieckmann, H.J.; Kim, K.K.; Yokota, M.; Kim, R.; Kim, S.H. Structure-based assignment of the biochemical function of a hypothetical protein: A test case of structural genomics. Proc. Natl. Acad. Sci. USA 1998, 95, 15189–15193.
- Peat, T.S.; Newman, J.; Waldo, G.S.; Berendzen, J.; Terwilliger, T.C. Structure of translation initiation factor 5A from Pyrobaculum aerophilum at 1.75 Å resolution. Structure 1998, 6, 1207–1214.
- Tong, Y.; Park, I.; Hong, B.S.; Nedyalkova, L.; Tempel, W.; Park, H.W. Crystal structure of human eIF5A1: Insight into functional similarity of human eIF5A1 and eIF5A2. Proteins Struct. Funct. Bioinforma. 2009, 75, 1040–1045.
- Cooper, H.L.; Park, M.H.; Folk, J.E. Posttranslational formation of hypusine in a single major protein occurs generally in growing cells and is associated with activation of lymphocyte growth. Cell 1982, 29, 791–797.
- Rossi, D.; Barbosa, N.M.; Galvão, F.C.; Boldrin, P.E.G.; Hershey, J.W.B.; Zanelli, C.F.; Fraser, C.S.; Valentini, S.R. Evidence for a negative cooperativity between eIF5A and eEF2 on binding to the ribosome. PLoS ONE 2016, 11, e0154205.
- Dias, C.A.O.; Gregio, A.P.B.; Rossi, D.; Galvão, F.C.; Watanabe, T.F.; Park, M.H.; Valentini, S.R.; Zanelli, C.F. EIF5A interacts functionally with eEF2. Amino Acids 2012, 42, 697–702.
- Schmidt, C.; Becker, T.; Heuer, A.; Braunger, K.; Shanmuganathan, V.; Pech, M.; Berninghausen, O.; Wilson, D.N.; Beckmann, R. Structure of the hypusinylated eukaryotic translation factor eIF-5A bound to the ribosome. Nucleic Acids Res. 2015, 44, 1944–1951.
- Shi, X.P.; Yin, K.C.; Zimolo, Z.A.; Stern, A.M.; Waxman, L. The subcellular distribution of eukaryotic translation initiation factor, eIF-5A, in cultured cells. Exp. Cell Res. 1996, 225, 348–356.
- Valentini, S.R.; Casolari, J.M.; Oliveira, C.C.; Silver, P.A.; McBride, A.E. Genetic interactions of yeast eukaryotic translation initiation factor 5a (eIF5A) reveal connections to poly(A)-binding protein and protein kinase C signaling. Genetics 2002, 160, 393–405.
- Rossi, D.; Galvão, F.C.; Bellato, H.M.; Boldrin, P.E.G.; Andrews, B.J.; Valentini, S.R.; Zanelli, C.F. eIF5A has a function in the cotranslational translocation of proteins into the ER. Amino Acids 2014, 46, 645–653.
- Barba-Aliaga, M.; Mena, A.; Espinoza, V.; Apostolova, N.; Costell, M.; Alepuz, P. Hypusinated eIF5A is required for the translation of collagen. J. Cell Sci. 2021, 134, jcs258643.
- Mandal, A.; Mandal, S.; Park, M.H. Global quantitative proteomics reveal up-regulation of endoplasmic reticulum stress response proteins upon depletion of eIF5A in HeLa cells. Sci. Rep. 2016, 6, 25795.
- Xu, A.; Chen, K.Y. Hypusine Is Required for a Sequence-specific Interaction of Eukaryotic Initiation Factor 5A with Postsystematic Evolution of Ligands by Exponential Enrichment RNA. J. Biol. Chem. 2001, 276, 2555–2561.
- Xu, A.; Jao, D.L.E.; Chen, K.Y. Identification of mRNA that binds to eukaryotic initiation factor 5A by affinity co-purification and differential display. Biochem. J. 2004, 384, 585–590.
- Aksu, M.; Trakhanov, S.; Görlich, D. Structure of the exportin Xpo4 in complex with RanGTP and the hypusine-containing translation factor eIF5A. Nat. Commun. 2016, 7, 11952.
- Zuk, D.; Jacobson, A. A single amino acid substitution in yeast eIF-5A results in mRNA stabilization. EMBO J. 1998, 17, 2914–2925.
- Schrader, R.; Young, C.; Kozian, D.; Hoffmann, R.; Lottspeich, F. Temperature-sensitive eIF5A mutant accumulates transcripts targeted to the nonsense-mediated decay pathway. J. Biol. Chem. 2006, 281, 35336–35346.
- Bassani, F.; Zink, I.A.; Pribasnig, T.; Wolfinger, M.T.; Romagnoli, A.; Resch, A.; Schleper, C.; Bläsi, U.; La Teana, A. Indications for a moonlighting function of translation factor aIF5A in the crenarchaeum Sulfolobus solfataricus. RNA Biol. 2019, 16, 675–685.
- Park, M.H.; Joe, Y.A.; Kang, K.R.; Lee, Y.B.; Wolff, E.C. The polyamine-derived amino acid hypusine: Its post-translational formation in eIF-5A and its role in cell proliferation. Amino Acids 1996, 10, 109–121.
- Hanauske-Abel, H.M.; Slowinska, B.; Zagulska, S.; Wilson, R.C.; Staiano-Coico, L.; Hanauske, A.R.; McCaffrey, T.; Szabo, P. Detection of a sub-set of polysomal mRNAs associated with modulation of hypusine formation at the G1-S boundary Proposal of a role for eIF-5A in onset of DNA replication. FEBS Lett. 1995, 366, 92–98.
- Chen, Z.P.; Yan, Y.P.; Ding, Q.J.; Knapp, S.; Potenza, J.A.; Schugar, H.J.; Chen, K.Y. Effects of inhibitors of deoxyhypusine synthase on the differentiation of mouse neuroblastoma and erythroleukemia cells. Cancer Lett. 1996, 105, 233–239.
- Clement, P.M.; Hanauske-Abel, H.M.; Wolff, E.C.; Kleinman, H.K.; Park, M.H. The antifungal drug ciclopirox inhibits deoxyhypusine and proline hydroxylation, endothelial cell growth and angiogenesis in vitro. Int. J. Cancer 2002, 100, 491–498.
- Nishimura, K.; Lee, S.B.; Park, J.H.; Park, M.H. Essential role of eIF5A-1 and deoxyhypusine synthase in mouse embryonic development. Amino Acids 2012, 42, 703–710.
- Sievert, H.; Pällmann, N.; Miller, K.K.; Hermans-Borgmeyer, I.; Venz, S.; Sendoel, A.; Preukschas, M.; Schweizer, M.; Boettcher, S.; Janiesch, P.C.; et al. A novel mouse model for inhibition of DOHH-mediated hypusine modification reveals a crucial function in embryonic development, proliferation and oncogenic transformation. DMM Dis. Model. Mech. 2014, 7, 963–976.
- Padgett, L.R.; Robertson, M.A.; Anderson-Baucum, E.K.; Connors, C.T.; Wu, W.; Mirmira, R.G.; Mastracci, T.L. Deoxyhypusine synthase, an essential enzyme for hypusine biosynthesis, is required for proper exocrine pancreas development. FASEB J. 2021, 35, e21473.
- Levasseur, E.M.; Yamada, K.; Piñeros, A.R.; Wu, W.; Syed, F.; Orr, K.S.; Anderson-Baucum, E.; Mastracci, T.L.; Maier, B.; Mosley, A.L.; et al. Hypusine biosynthesis in β cells links polyamine metabolism to facultative cellular proliferation to maintain glucose homeostasis. Sci. Signal. 2019, 12.
- Lubas, M.; Harder, L.M.; Kumsta, C.; Tiessen, I.; Hansen, M.; Andersen, J.S.; Lund, A.H.; Frankel, L.B. eIF 5A is required for autophagy by mediating ATG 3 translation. EMBO Rep. 2018, 19, e46072.
- Zhang, H.; Alsaleh, G.; Feltham, J.; Sun, Y.; Napolitano, G.; Riffelmacher, T.; Charles, P.; Frau, L.; Hublitz, P.; Yu, Z.; et al. Polyamines Control eIF5A Hypusination, TFEB Translation, and Autophagy to Reverse B Cell Senescence. Mol. Cell 2019, 76, 110–125.
- Zanelli, C.F.; Valentini, S.R. Pkc1 acts through Zds1 and Gic1 to suppress growth and cell polarity defects of a yeast eIF5A mutant. Genetics 2005, 171, 1571–1581.
- Chatterjee, I.; Gross, S.R.; Kinzy, T.G.; Chen, K.Y. Rapid depletion of mutant eukaryotic initiation factor 5A at restrictive temperature reveals connections to actin cytoskeleton and cell cycle progression. Mol. Genet. Genomics 2006, 275, 264–276.
- Nguyen, S.; Leija, C.; Kinch, L.; Regmi, S.; Li, Q.; Grishin, N.V.; Phillips, M.A. Deoxyhypusine modification of eukaryotic translation initiation factor 5A (eIF5A) is essential for Trypanosoma brucei growth and for expression of polyprolyl-containing proteins. J. Biol. Chem. 2015, 290, 19987–19988.
- Li, T.; Belda-Palazón, B.; Ferrando, A.; Alepuz, P. Fertility and polarized cell growth depends on eIF5A for translation of polyproline-rich formins in Saccharomyces cerevisiae. Genetics 2014, 197, 1191–1200.
- Muñoz-Soriano, V.; Domingo-Muelas, A.; Li, T.; Gamero, E.; Bizy, A.; Fariñas, I.; Alepuz, P.; Paricio, N. Evolutionary conserved role of eukaryotic translation factor eIF5A in the regulation of actin-nucleating formins. Sci. Rep. 2017, 7, 9580.
- Fujimura, K.; Choi, S.; Wyse, M.; Strnadel, J.; Wright, T.; Klemke, R. Eukaryotic translation initiation factor 5A (EIF5A) regulates pancreatic cancer metastasis by modulating RhoA and rho-associated kinase (ROCK) protein expression levels. J. Biol. Chem. 2015, 290, 29907–29919.
- Coni, S.; Serrao, S.M.; Yurtsever, Z.N.; Di Magno, L.; Bordone, R.; Bertani, C.; Licursi, V.; Ianniello, Z.; Infante, P.; Moretti, M.; et al. Blockade of EIF5A hypusination limits colorectal cancer growth by inhibiting MYC elongation. Cell Death Dis. 2020, 11, 1045.
- Ning, L.; Wang, L.; Zhang, H.; Jiao, X.; Chen, D. Eukaryotic translation initiation factor 5A in the pathogenesis of cancers (Review). Oncol. Lett. 2020, 20, 81.
- Li, A.L.; Li, H.Y.; Jin, B.F.; Ye, Q.N.; Zhou, T.; Yu, X.D.; Pan, X.; Man, J.H.; He, K.; Yu, M.; et al. A novel eIF5A complex functions as a regulator of p53 and p53-dependent apoptosis. J. Biol. Chem. 2004, 279, 49251–49258.
- Caraglia, M.; Park, M.H.; Wolff, E.C.; Marra, M.; Abbruzzese, A. EIF5A isoforms and cancer: Two brothers for two functions? Amino Acids 2013, 44, 103–109.
- Tan, X.; Wang, D.B.; Lu, X.; Wei, H.; Zhu, R.; Zhu, S.S.; Jiang, H.; Yang, Z.J. Doxorubicin induces apoptosis in H9c2 cardiomyocytes: Role of overexpressed Eukaryotic translation initiation factor 5A. Biol. Pharm. Bull. 2010, 33, 1666–1672.
- Martella, M.; Catalanotto, C.; Talora, C.; La Teana, A.; Londei, P.; Benelli, D. Inhibition of eukaryotic translation initiation factor 5a (Eif5a) hypusination suppress p53 translation and alters the association of eif5a to the ribosomes. Int. J. Mol. Sci. 2020, 21, 4583.
- Ruhl, M.; Himmelspach, M.; Bahr, G.M.; Hammerschmid, F.; Jaksche, H.; Wolff, B.; Aschauer, H.; Farrington, G.K.; Probst, H.; Bevec, D.; et al. Eukaryotic initiation factor 5A is a cellular target of the human immunodeficiency virus type 1 Rev activation domain mediating trans- activation. J. Cell Biol. 1993, 123, 1309–1320.
- Rosorius, O.; Reichart, B.; Krätzer, F.; Heger, P.; Dabauvalle, M.C.; Hauber, J. Nuclear pore localization and nucleocytoplasmic transport of eIF-5A: Evidence for direct interaction with the export receptor CRM1. J. Cell Sci. 1999, 112, 2369–2380.
- Olsen, M.E.; Connor, J.H. Hypusination of eIF5A as a Target for Antiviral Therapy. DNA Cell Biol. 2017, 36, 198–201.
- Tersey, S.A.; Colvin, S.C.; Maier, B.; Mirmira, R.G. Protective effects of polyamine depletion in mouse models of type 1 diabetes: Implications for therapy. Amino Acids 2014, 46, 633–642.
- Maier, B.; Ogihara, T.; Trace, A.P.; Tersey, S.A.; Robbins, R.D.; Chakrabarti, S.K.; Nunemaker, C.S.; Stull, N.D.; Taylor, C.A.; Thompson, J.E.; et al. The unique hypusine modification of eIF5A promotes islet β cell inflammation and dysfunction in mice. J. Clin. Investig. 2010, 120, 2156–2170.
- Jenkins, Z.A.; Hååg, P.G.; Johansson, H.E. Human EIF5A2 on chromosome 3q25-q27 is a phylogenetically conserved vertebrate variant of eukaryotic translation initiation factor 5A with tissue-specific expression. Genomics 2001, 71, 101–109.
- Clement, P.M.J.; Henderson, C.A.; Jenkins, Z.A.; Smit-McBride, Z.; Wolff, E.C.; Hershey, J.W.B.; Park, M.H.; Johansson, H.E. Identification and characterization of eukaryotic initiation factor 5A-2. Eur. J. Biochem. 2003, 270, 4254–4263.
- Park, M.H.; Kar, R.K.; Banka, S.; Ziegler, A.; Chung, W.K. Post-translational formation of hypusine in eIF5A: Implications in human neurodevelopment. Amino Acids 2021, 1–15.
- Hyvönen, M.T.; Keinänen, T.A.; Khomutov, M.; Simonian, A.; Vepsäläinen, J.; Park, J.H.; Khomutov, A.R.; Alhonen, L.; Park, M.H. Effects of novel C-methylated spermidine analogs on cell growth via hypusination of eukaryotic translation initiation factor 5A. Amino Acids 2012, 42, 685–695.
- Mounce, B.C.; Cesaro, T.; Moratorio, G.; Hooikaas, P.J.; Yakovleva, A.; Werneke, S.W.; Smith, E.C.; Poirier, E.Z.; Simon-Loriere, E.; Prot, M.; et al. Inhibition of Polyamine Biosynthesis Is a Broad-Spectrum Strategy against RNA Viruses. J. Virol. 2016, 90, 9683–9692.
- Li, J.; Meng, Y.; Wu, X.; Sun, Y. Polyamines and related signaling pathways in cancer. Cancer Cell Int. 2020, 20, 1–16.
- Wolff, E.C.; Kang, K.R.; Kim, Y.S.; Park, M.H. Posttranslational synthesis of hypusine: Evolutionary progression and specificity of the hypusine modification. Amino Acids 2007, 33, 341–350.
- Lou, B.; Fan, J.; Wang, K.; Chen, W.; Zhou, X.; Zhang, J.; Lin, S.; Lv, F.; Chen, Y. N1-guanyl-1,7-diaminoheptane (GC7) enhances the therapeutic efficacy of doxorubicin by inhibiting activation of eukaryotic translation initiation factor 5A2 (eIF5A2) and preventing the epithelial-mesenchymal transition in hepatocellular carcinoma cells. Exp. Cell Res. 2013, 319, 2708–2717.
- Bandino, A.; Geerts, D.; Koster, J.; Bachmann, A.S. Deoxyhypusine synthase (DHPS) inhibitor GC7 induces p21/Rb-mediated inhibition of tumor cell growth and DHPS expression correlates with poor prognosis in neuroblastoma patients. Cell. Oncol. 2014, 37, 387–398.
- Guerville, F.; De Souto Barreto, P.; Ader, I.; Andrieu, S.; Casteilla, L.; Dray, C.; Fazilleau, N.; Guyonnet, S.; Langin, D.; Liblau, R.; et al. Revisiting the Hallmarks of Aging to Identify Markers of Biological Age. J. Prev. Alzheimer’s Dis. 2020, 7, 56–64.
- Lee, H.; Smith, S.B.; Yoon, Y. The short variant of the mitochondrial dynamin OPA1 maintains mitochondrial energetics and cristae structure. J. Biol. Chem. 2017, 292, 7115–7130.
- Arlt, H.; Steglich, G.; Perryman, R.; Guiard, B.; Neupert, W.; Langer, T. The formation of respiratory chain complexes in mitochondria is under the proteolytic control of the m-AAA protease. EMBO J. 1998, 17, 4837–4847.
- Chen, Y.C.; Cheng, T.H.; Lin, W.L.; Chen, C.L.; Yang, W.Y.; Blackstone, C.; Chang, C.R. Srv2 Is a Pro-fission Factor that Modulates Yeast Mitochondrial Morphology and Respiration by Regulating Actin Assembly. iScience 2019, 11, 305–317.
- Hansen, K.G.; Herrmann, J.M. Transport of Proteins into Mitochondria. Protein J. 2019, 38, 330–342.
- Hersch, S.J.; Elgamal, S.; Katz, A.; Ibba, M.; Navarre, W.W. Translation initiation rate determines the impact of ribosome stalling on bacterial protein synthesis. J. Biol. Chem. 2014, 289, 28160–28171.
- Turpaev, K.T. Translation Factor eIF5A, Modification with Hypusine and Role in Regulation of Gene Expression. eIF5A as a Target for Pharmacological Interventions. Biochemistry 2018, 83, 863–873.
- Sun, Z.; Cheng, Z.; Taylor, C.A.; Mcconkey, B.J.; Thompson, J.E. Apoptosis induction by eIF5A1 involves activation of the intrinsic mitochondrial pathway. J. Cell. Physiol. 2010, 223, 798–809.
- Pereira, K.D.; Tamborlin, L.; de Lima, T.I.; Consonni, S.R.; Silveira, L.R.; Luchessi, A.D. Alternative human eIF5A protein isoform plays a critical role in mitochondria. J. Cell. Biochem. 2021, 122, 549–561.
- Grancara, S.; Dalla Via, L.; García-Argáez, A.N.; Ohkubo, S.; Pacella, E.; Manente, S.; Bragadin, M.; Toninello, A.; Agostinelli, E. Spermine cycling in mitochondria is mediated by adenine nucleotide translocase activity: Mechanism and pathophysiological implications. Amino Acids 2016, 48, 2327–2337.
- Miyake, T.; Pradeep, S.; Wu, S.Y.; Rupaimoole, R.; Zand, B.; Wen, Y.; Gharpure, K.M.; Nagaraja, A.S.; Hu, W.; Cho, M.S.; et al. XPO1/CRM1 inhibition causes antitumor effects by mitochondrial accumulation of eIF5A. Clin. Cancer Res. 2015, 21, 3286–3297.
- Liu, J.; Zhan, X.; Li, M.; Li, G.; Zhang, P.; Xiao, Z.; Shao, M.; Peng, F.; Hu, R.; Chen, Z. Mitochondrial proteomics of nasopharyngeal carcinoma metastasis. BMC Med. Genomics 2012, 5, 1–17.
- Pereira, K.D.; Tamborlin, L.; Meneguello, L.; de Proença, A.R.G.; Almeida, I.C.D.P.A.; Lourenço, R.F.; Luchessi, A.D. Alternative Start Codon Connects eIF5A to Mitochondria. J. Cell. Physiol. 2016, 231, 2682–2689.
- Melis, N.; Rubera, I.; Cougnon, M.; Giraud, S.; Mograbi, B.; Belaid, A.; Pisani, D.F.; Huber, S.M.; Lacas-Gervais, S.; Fragaki, K.; et al. Targeting eIF5A hypusination prevents anoxic cell death through mitochondrial silencing and improves kidney transplant outcome. J. Am. Soc. Nephrol. 2017, 28, 811–822.
- Puleston, D.J.; Buck, M.D.; Klein Geltink, R.I.; Kyle, R.L.; Caputa, G.; O’Sullivan, D.; Cameron, A.M.; Castoldi, A.; Musa, Y.; Kabat, A.M.; et al. Polyamines and eIF5A Hypusination Modulate Mitochondrial Respiration and Macrophage Activation. Cell Metab. 2019, 30, 1–12.
- Hofer, S.J.; Liang, Y.T.; Zimmermann, A.; Schroeder, S.; Dengjel, J.; Kroemer, G.; Eisenberg, T.; Sigrist, S.J.; Madeo, F. Spermidine-induced hypusination preserves mitochondrial and cognitive function during aging. Autophagy 2021, 17, 2037–2039.
