Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Virology
CMV is a major cause of morbidity and mortality in immunocompromised individuals that will benefit from the availability of a vaccine. Despite the efforts made during the last decade, no CMV vaccine is available. An ideal CMV vaccine should elicit a broad immune response against multiple viral antigens including proteins involved in virus-cell interaction and entry. However, the therapeutic use of neutralizing antibodies targeting glycoproteins involved in viral entry achieved only partial protection against infection.
- cytomegalovirus
- pangenome
- proteome
1. Introduction
Human cytomegalovirus (CMV) is a large envelope worldwide prevalent betaherpesvirus, ranging from 45% to 100% in the general population based on socio-economic factors [1,2]. Although CMV generally causes asymptomatic infections in immunocompetent individuals, it is a major cause of morbidity and mortality in immunocompromised individuals such as organ transplant recipients, AIDS, and with congenital infection [3,4,5,6,7].
The CMV genome (236 kb) consists of a unique long (UL) and a unique short (US) region flanked by inverted repeats. CMV gene expression occurs with the expression of immediate-early genes followed by early, early-late and late transcripts [8]. In addition to the 165 canonical ORFs [9,10], CMV genome encodes for other alternative transcripts in addition to have non-canonical translation initiation sites [11,12]. Furthermore, CMV encodes for a large number of genes, many of them with unknown functions that may probably be involved in key processes during host-cell interaction [13].
CMV is able to infect a high number of cell types including fibroblasts, endothelial cells, epithelial cells and myeloid lineage cells, among others [14,15]. CMV is a highly complex virus with multiple proteins embedded in the viral envelope, with at least four distinct types of covalently linked glycoprotein complexes required for CMV infectivity including gCI complex (gB dimer), gCII complex (gM, gN), gCIII complex (trimer gH, gL, gO) and pentameric complex (gH/gL/UL128-131) [16,17]. The therapeutic use of neutralizing antibodies, targeting glycoproteins mediating viral entry, have demonstrated to only achieve partial protection against CMV infection [18,19,20,21,22,23]. One possible explanation if that other proteins may be involved in viral entry that might be also necessary to target in order to elicit a complete protection against infection. An ideal vaccine against CMV infection should elicit a broad immune response, including both neutralizing antibody and T-cell response, against multiple viral antigens including proteins involved in virus-cell interaction and entry [24,25], which may increase efficacy compared with the previously tested vaccines [1,26,27,28]. In fact, despite the efforts made during the last decade, no CMV vaccine is still available [18,27,29,30]. Thus, understanding the complete repertoire of CMV proteins involved in cell entry may also help to determine the neutralizing response necessary to block infection and may provide novel candidates that could be included in new vaccines design.
2. Identification of Putative Transmembrane Proteins
To determine the transmembrane regions of the proteins encoded by the CMV genome, the genome of nine different CMV strains including both clinical and laboratory strains (Table 1) were analyzed using three different bioinformatic methods: Phobius, PureseqTM and TMHMM. A description of the methodological approach used is represented in Figure 1. CMV is known to accumulate mutations quite rapidly in cell culture during cell passaging [31]. In order to test to what level these nine selected CMV strains are representative of the 335 available CMV genomes in GenBank, the 56190 ORFs were aligned with the ORFs in our CMV dataset. We obtained 100% median percentage identity and breadth coverage (overlapping distance), representing 99.95% of the total ORFs from the Human betaherpesvirus 5 in the NCBI database.
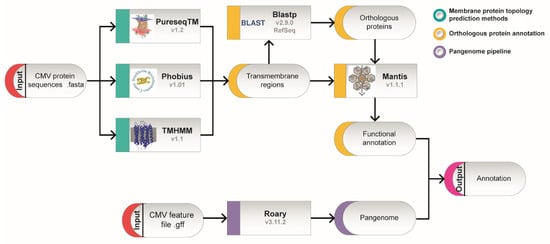
Figure 1. Schematic representation of the applied workflow. Fasta format protein sequences from nine CMV genomes were analyzed in parallel to predict transmembrane domains and to create an entire set of genes from all strains (pangenome). Transmembrane topology was studied following three different approaches: PureseqTM, Phobius and TMHMM, under default parameters. Predicted transmembrane proteins were compared with orthologous proteins identified by BLAST with the whole Mantis database for the prediction of functional annotation. Proteins that were common to all nine genome datasets formed the core protein set, and functions were annotated accordingly for each transmembrane protein.
Table 1. Characteristics of CMV strains used in this study and their corresponding accession number at nucleotide database.
| CMV Strains | Isolation Source | Number of Culture Passages | Accession Number |
|---|---|---|---|
| AD169 | Adenoids of a 7-year-old girl | Many times in human fibroblasts | FJ527563.1 |
| Towne | Urine of a 2-month-old infant with microcephaly and hepatosplenomegaly | Many times in human fibroblasts | FJ616285.1 |
| Toledo | Urine from a congenitally infected infant | Several times in human fibroblasts | GU937742.2 |
| TR | Vitreous humor from eye of HIV-positive male | Several times in human fibroblasts | KF021605.1 |
| VR7863 | Urine samples of a congenitally infected neonate and cultured in endothelial and epithelial cells | Cultured in endothelial and epithelial cells | KX544838.1 |
| TB40-E_UNC | Throat swab of a bone marrow transplant patient | Cultured adapted | KX544839.1 |
| HANSCTR4 | Blood from stem cell transplant recipient (D-R+) | Sequenced directly from clinical material via target enrichment | KY123653.1 |
| AD169-BAC20 | - | - | MN920393.1 |
| Merlin | Urine from a congenitally infected child | 3 times in human fibroblasts | NC006273.2 |
Based on the first analysis, we identified 94 proteins with potential transmembrane domains (Figure 2). Seventeen of them were not considered for further analysis because of the following reasons. Proteins UL74, UL115, UL47, UL49, UL76, US22, UL77, UL105, UL122 and UL89 were previously described not to be part of membrane structures [32,33,34,35,36,37,38,39,40,41]. UL47, UL49, UL76 and US22 proteins are known to be part of the tegument, UL77 is located in the capsid; UL105, UL122 and UL89 are found in the nucleus of the host cells [32,33,34,35,36,37,38,39]. In addition, UL4, UL22A and UL116, which were predicted to have one transmembrane domain, were discarded because the transmembrane domain corresponded to the sequence of signal peptide [42,43,44]. In addition, RL13TRL14 and US33 (TB40-E_UNC strain) or ORFL27C and ORFL49W.IORF1 (AD169-BAC20) proteins were only found in one of the CMV studied strains and were not considered for further analysis.
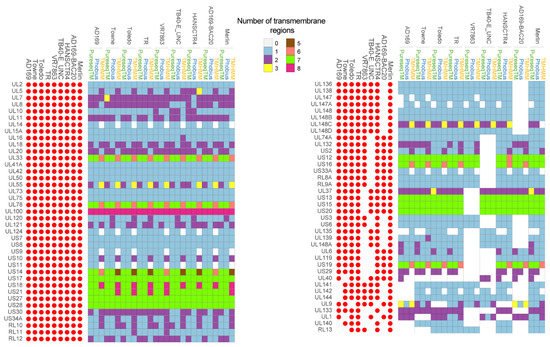
Figure 2. Predicted transmembrane proteins for the studied CMV genomes. Proteins with at least one predicted transmembrane domain with one of three tested methods were annotated as transmembrane proteins. The number of transmembrane domains for each protein was represented using the indicated chromatic scale ranging from zero to eight regions for each of the three methods used (PureseqTM, Phobius and TMHMM). For each strain, the presence of the gene was represented with the filled red circles and absent genes with empty red circles.
For further characterization of the 77 remaining proteins with predicted transmembrane (TM) domains, a systematic review was performed to search for any previous published information. A graphical representation of the number of predicted TM regions found for each ORF, in each of the nine strains with the indicated bioinformatics tool is shown in Figure 2. Of the 77 proteins analyzed, 33 (43%) proteins only had one TM domain, 23 (29.87%) had from 1 to 2 TM domains, 6 (7.7%) exhibited 1-3, while 15 proteins (19.48%) had from 5 to 8 TM domains. None of the analyzed proteins had four TM domains.
Nineteen out of the 77 proteins (UL2, UL6, UL9, UL14, UL15A, UL74A, UL120, UL121, UL140, UL148C, UL148D, US13, US15, US19, US29, US30, US33A, RL8A, RL9A and RL10) have no previously described function, 13 (UL1, UL5, UL8, UL10, UL20, UL42, UL78, UL124, UL139, UL147, US34A, RL12 and RL13) have been partially studied, 1 (UL41A) has previously been shown not to code for a protein [10] and the other 43 proteins have a previously described function (Table 2).
Table 2. CMV predicted transmembrane proteins indicating the cellular localization based on biotool Uniprop, the ascribed functions based on a bibliographic search and the number of predicted domains using the three different tools. (*) indicates unknown or non-verified function.
| Gene | Localization | Function | Number of TM Domains | References |
|---|---|---|---|---|
| UL1 * | VM | Unknown. pUL1 could modulate CMV host cell tropism. | 1–2 | [45] |
| UL2 * | HM | Unknown. | 1 | - |
| UL5 * | V | Unknown. It is suggested to be involved in efficient viral assembly, propagation and replication. | 1–3 | [46,47] |
| UL6 * | HM | Unknown. | 1–2 | - |
| UL7 | UL7 is involved in immunomodulation. | 2–3 | [48,49,50] | |
| UL8 * | HM | UL8 decreases the release of a large number of pro-inflammatory factors later after infection of THP-1 myeloid cells. UL8 may exert an immunosuppressive role key for CMV survival in the host. | 1–2 | [51] |
| UL9 * | HM | Unknown function. Its deletion mutation cause enhanced growth in HFFs cells. |
1–3 | [9] |
| UL10 * | M | Unknown. Potential role in immunomodulation. | 1–2 | [52] |
| UL11 | HM, ERM | pUL11 interacts with CD45 phosphatase on T cells, inducing the IL-10 secretion. | 1–2 | [53] |
| UL14 * | HM | Unknown. | 0–1 | - |
| UL15A * | HM | Unknown. | 1 | - |
| UL16 | HM | Immunoevasion and inhibition of the activation of NK cells. | 1 | [54] |
| UL18 | HM | Immunomodulation and immunoevasion. | 1–2 | [55] |
| UL20 * | ERM | Unknown. UL20 could be destined to sequester cellular proteinases not known to date for degradation in lysosomes. | 1–2 | [56] |
| UL33 | HM | UL33 has homology with GPCR which activates different ligand-independent signalling pathways and also involved in virus dissemination. | 6–7 | [57,58,59] |
| UL37 | ERM, GM, MM | Viral replication. | 2–3 | [60,61] |
| UL40 | HM | Immunomodulation. | 0–2 | [62] |
| UL41A * | VM | Unknown. UL41A not to code for proteins. | 1 | [10] |
| UL42 * | HM, C | Unknown. Potential role in immunoevasion. | 1 | [63,64] |
| UL50 | HNM | Assembly, maturation and egress of virions. | 1 | [65] |
| UL55 | VM, HM, GM | Glycoprotein B participates in viral entry. | 1–3 | [66] |
| UL73 | VM, HM, GM | Glycoprotein N is involved in the binding of the virus to the host cell, viral spread and virion morphogenesis. | 1 | [67] |
| UL74A * | VM | Unknown | 1 | - |
| UL75 | HM, VM | Glycoprotein H participates in viral entry. It is part of the trimeric and pentameric complexes. |
1 | [68] |
| UL78 * | HM, ERM | Unknown. UL78 is a G protein-coupled receptor. | 6–7 | [69,70] |
| UL100 | HM, VM | Envelope glycoprotein M participates in viral entry. | 8 | [71,72] |
| UL119 | VM | Immunoevasion. | 1 | [73] |
| UL120 * | HM | Unknown. | 1–2 | - |
| UL121 * | HM | Unknown. | 1–2 | - |
| UL124 * | HM | Potential role in latency. | 0–1 | [74] |
| UL132 | VM | Essential for CMV assembly compartment formation and the efficient production of infectious particles. | 1–2 | [75] |
| UL133 | GM | UL133 forms a complex with UL138 and UL136. It is involved in the establishment of CMV latency. | 2 | [76] |
| UL135 | HM, GM | Immunomodulation. Post entry Tropism in Endothelial Cells. | 0–1 | [77,78] |
| UL136 | HM | Replication, latency, and dissemination. Post entry Tropism in Endothelial Cells. | 1 | [76,78,79,80] |
| UL138 | GM | Latency and DNA replication. | 1 | [81,82] |
| UL139 * | HM | Unknown. Potential role in immunomodulation. | 1–2 | [83] |
| UL140 * | HM | Unknown. | 1 | - |
| UL141 | ERM | Immunomodulation and DNA replication. | 1 | [84,85,86] |
| UL142 | ERM | Immunomodulation. | 0–1 | [87] |
| UL144 | HM | Inhibition of T-cell activation and latency. | 1 | [88,89] |
| UL147 * | EXR | Unknown. Potential role in immunomodulation. | 0–1 | [90] |
| UL147A | HM | Immunomodulation. | 0–1 | [91] |
| UL148 | ERM | Viral ER-resident glycoprotein that interacts with UL116 promoting the incorporation of gH/gL complexes into virions. | 1 | [92] |
| UL148A | HM | Immunoevasion of NK cells. | 1–2 | [93] |
| UL148B * | HM | Unknown. | 1 | [94] |
| UL148C * | HM | Unknown. | 0–3 | [94] |
| UL148D * | HM | Unknown. | 1 | [94] |
| US2 | ERM | Immunomodulation. | 1–2 | [95] |
| US3 | ERM | Immunoevasion. | 1 | [96] |
| US6 | ERM | Immunomodulation. | 1 | [97] |
| US7 | ERM | Immunoevasion. | 1 | [98] |
| US8 | ERM, GM | Immunomodulation. | 1 | [98] |
| US9 | ERM, GM, CK | Glycoprotein US9 is an antagonist of IFN signalling to persistently evade host innate antiviral responses. | 0–1 | [99] |
| US10 | ERM | Inhibition of the host immune response. | 1–2 | [100] |
| US11 | ERM | Inhibition of the host immune response. | 0–1 | [101] |
| US12 | HM | Inmunomodulation of NK cells activation. | 6–7 | [102] |
| US13 * | HM | Unknown. | 7 | - |
| US14 | HM | Inmunomodulation of NK cells activation. Potential role in virions maturation and egress. | 5–7 | [102,103] |
| US15 * | HM | Unknown. | 7 | - |
| US16 | HM, C | Tropism in endothelial and epithelial cells. | 6–7 | [104] |
| US17 | HM | Immunomodulation. | 7 | [105] |
| US18 | HM. | Immunoevasion of NK cell. | 7–8 | [106] |
| US19 * | HM | Unknown. Its delection affect NK cell activation. | 6–7 | [102] |
| US20 | M | Inhibition NK cell activation. Also participates in the viral replication process in endothelial cells. | 7 | [106,107] |
| US21 | HM | Viroporin that modulates calcium homeostasis and protects cells against apoptosis. | 7–8 | [108] |
| US27 | V, HM | Immunomodulation. Also is required for efficient viral spread by the extracellular route. | 7 | [109,110,111] |
| US28 | HM | Immunomodulation. Lytic and latent CMV infection. Possible role in regulation of the actin cytoskeleton or cytoskeletal remodelling. | 7 | [112,113] |
| US29 * | HM | Unknown. | 0–2 | - |
| US30 * | HM | Unknown. | 1–2 | - |
| US33A * | - | Unknown. | 0–1 | [114] |
| US34A * | HM. | Unknown. Potential target of SUMO complex. | 1–2 | [115] |
| RL8A * | HM | Unknown. | 1 | - |
| RL9A * | HM | Unknown. | 1 | - |
| RL10 * | VM | Unknown. | 1–2 | - |
| RL11 | HM | Immunomodulation. RL11 is a type I transmembrane glycoproteins which bind immunoglobulin G Fc. I | 1–2 | [116] |
| RL12 * | VM | Unknown. RL12 is a Fc binding protein. | 1–2 | [117] |
| RL13 * | VM | Unknown. Potential role in replication, immunoevasión and viral spread by cell-free or cell-to-cell mechanisms. | 1 | [118,119,120] |
* indicates unknown or non-verified function. CK: Cytoskeleton C: Cytoplasm, ERM: Host endoplasmic reticulum membrane, EXR: Extracellular region, GM: Golgi reticulum membrane, HM: host membrane, HMN: Host nucleus membrane, M: Membrane, MM: Mitochondrion membrane, V: Virion, VM: Virion membrane.
The number of predicted domains differed in some of the studied proteins when using different methods. The results obtained TMHMM method were the most divergent of the three tested methods. On the contrary, a group of proteins encoded by the genes UL33, UL78 and the genes from the unique short (US) region US12-US21, US27 and US28 proteins were predicted to have more than five TM regions by all three methods. In fact, TM regions of these genes, such as the members of US12 family and the proteins with homology to the chemokine receptor family of G protein-coupled receptors (GPCRs): US27 and US28, have been previously described supporting our results [108,109,121].
A validation experiment was carried out using as an example UL2 and UL124, two of the identified proteins with unknown function. The ORF encoding for these two proteins were cloned into a eukaryotic expression plasmid that included a Myc tag sequence in the 5´end of the clone products. After transfecting the HEK 293T mammalian cell line, plasma membrane proteins were extracted and the cytoplasmic (C) and plasma membrane (PM) protein fractions were tested by Western Blot using an anti Myc antibody.
3. Homology Analysis of thePredicted Transmembrane Proteins
In addition to the exhaustive systematic review of the literature, further analysis of sequence homology with known proteins from other organisms was performed using Mantis software. Based on this analysis, we found homologies for two of the proteins with unknown function. UL139 had some level of homology (e-value = 5.1 × 10−28) with proteins involved in cell adhesion, while UL15A had some homology (e value = 1.53 × 10−4) with a biotin permease protein. UL15A ORF was identified in all 9 CMV strains analyzed, while UL139 that was only present in the TR strain.
In addition, Mantis analysis shed an association of UL1 with a carcinoembryonic antigen-related cell adhesion molecule, which is a cell adhesion receptor of the immunoglobulin-like superfamily. UL78 was also identified by Mantis as seven transmembrane receptor from the rhodopsin family. UL147 has been proposed by Mantis to be involved in immune response and chemokine activity and US33A seems to have a von Willebrand A (VWA) domain. However, US33A was present exclusively in Towne, Toledo, TR and VR7863 strains.
4. Sequence Differences among Strains
The analysis of the generated pangenome (Figure 2 and Figure 3), revealed wide differences among strains [13]. Clinical isolate VR7863 and TB40-E_UNC strains lacked a large number of genes compared to the other strains. The VR7863 strain lacked some genes with unknown functions such as UL1, UL6, UL139, UL140, US13, US15, US19 and US29 and other genes involved in immunoevasion, DNA packaging, latency or viral replication such as UL37, UL40, UL119, UL133, UL135, UL148A and US20 [39,60,61,62,73,76,77,93,106]. The TB40-E_UNC strain lacked some genes with unknown function such as UL6, UL74A, UL140, US12, US19, US29, US33A, RL8A, RL9A and RL13 and other genes involved in host immune response evasion, CMV assembly, tropism, or latency such as UL119, UL132, UL133, US2, US3 and US16 [73,75,95,96,102,104,118,119].
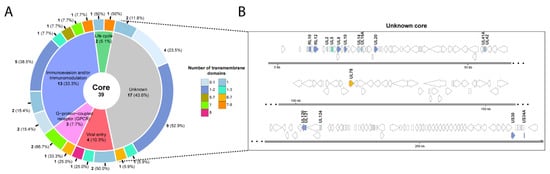
Figure 3. Functional analysis of the 39 core proteins. (A) Pie chart of the proteins found in all the studied strains were grouped based on their functions. For each group, the number of predicted transmembrane domains is also indicated. When the number of transmembrane domains predicted was different using the three methods, a range of values is shown. The number of proteins in each section is marked in blue and the percentage between brackets. (B) Genomic location of the 17 proteins with non-described function.
AD169 strain have a deletion of a genomic region that included UL140, UL141, UL142, UL144 genes and RL13 gene (known to have TM domains) [117]. All of them have functions related to the evasion of the host’s immune system [84,85,86,87,88]. In addition, the AD169 BACmid (Table 1) widely used for research, lacked several genes encoding TM proteins [117]. Some of them such as UL140, UL141, UL142, UL144 and RL13 were also deleted in AD169 strain. While the AD169 BACmid also lacked other genes such as UL135, UL136, UL138, UL148 and US3-US6 genes, involved in DNA replication, latency, virulence, tropism, evasion of the immune response and other genes such as UL139, UL147, UL148B, UL148C and UL148D with uncharacterized function [77,79,81,92,96,97,117]. The Toledo strain lacked RL13, UL9 and UL128 genes, while the Towne strain lacked RL13, UL1 and UL40 genes. The TR and Merlin strains, widely used in research included all the analyzed genes.
This entry is adapted from the peer-reviewed paper 10.3390/ijms23052768
This entry is offline, you can click here to edit this entry!
