Penicillium digitatum is a widespread pathogen responsible for the postharvest decay of citrus, one of the most economically important crops worldwide. Currently, chemical fungicides are still the main strategy to control the green mould disease caused by the fungus. In this scenario, understanding the molecular determinants underlying P. digitatum’s response to biological and chemical antifungals may help in the development of safer and more effective non-chemical control methods.
- Penicillium digitatum
- proteomics
- alpha-sarcin
- Beetin 27 (BE27)
1. Introduction
2. Deciphering Molecular Determinants Underlying Penicillium digitatum’s Response to Biological and Chemical Antifungal Agents by Tandem Mass Tag (TMT)-Based High-Resolution LC-MS/MS
In this work, the molecular determinants of Penicillium digitatum response to α-sarcin and BE27, two inhibitors of protein synthesis, were investigated. We further evaluated protein changes following treatment with the commonly used fungicide thiabendazole (TBZ), a microtubule-destabilizing drug that inhibits mitosis [20] with the aim to compare P. digitatum response to biological and chemical fungicides. An advanced quantitative proteomic approach based on Tandem Mass Tag (TMT) isobaric labelling and nano-liquid chromatography coupled with high resolution tandem mass spectrometry (nanoLC MS/MS) was used to identify and quantify the expression levels of proteins from treated and untreated P. digitatum mycelia (Figure 1).
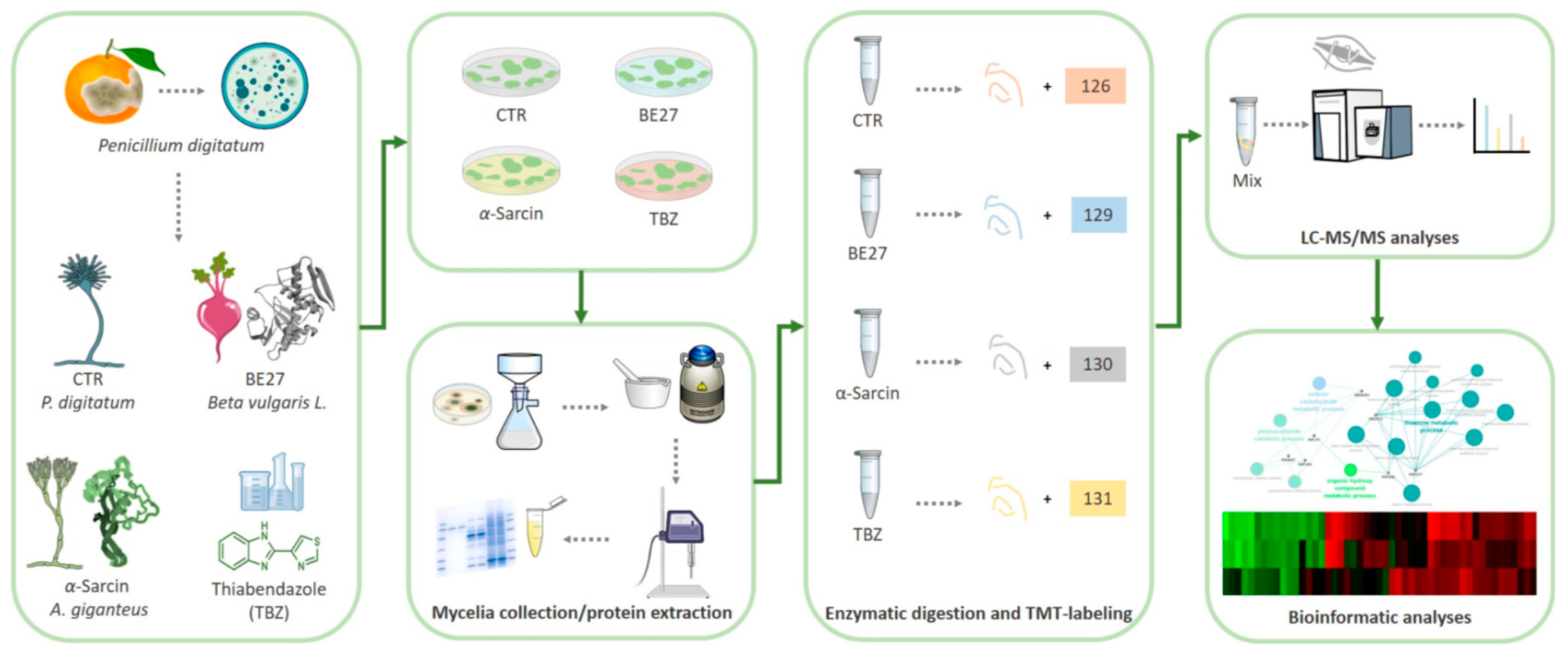
Figure 1. Schematic workflow applied for proteomic analysis of P. digitatum treated with fungicide compounds (i.e., α-sarcin, BE27 and TBZ).
Collectively, we identified candidate proteins potentially associated with α-sarcin, BE27 and TBZ treatment outcomes in P. digitatum. These were mainly involved in cell wall degradation and fungal morphogenesis, stress response, antioxidant and detoxification mechanisms and metabolic pathways. Although a similar trend in protein changes was driven by the three antifungal treatments, a distinct regulation in response to α-sarcin and BE27 treatments was also observed for a subset of proteins.
We focused primarily on differentially expressed proteins potentially impairing P. digitatum growth and virulence such as cell wall-degrading enzymes (CWDEs) and proteases. In phytopathogenic fungi, an arsenal of catalytic proteins supports nutrient acquisition, conidial formation, substrate colonization and host invasion [21]. We found several CWDEs and proteases that were differentially regulated upon protein toxins and TBZ antifungal treatments such as pectinesterases, pectate lyases, polygalacturonases, glucanases and peptidases. The importance of CWDEs in the virulence and pathogenesis of P. digitatum has been reported during the infection of oranges [22] and on postharvest citrus [23].
This entry is adapted from the peer-reviewed paper 10.3390/ijms23020680
References
- I. Talibi; H. Boubaker; E.H. Boudyach; A. Ait Ben Aoumar; Alternative methods for the control of postharvest citrus diseases. Journal of Applied Microbiology 2014, 117, 1-17, 10.1111/jam.12495.
- S. Tian; R. Torres; A-R. Ballester; B. Li; L. Vilanova; L. González-Candelas; Molecular aspects in pathogen-fruit interactions: Virulence and resistance. Postharvest Biology and Technology 2016, 122, 11-21, 10.1016/j.postharvbio.2016.04.018.
- Yulin Cheng; Yunlong Lin; Haohao Cao; Zhengguo Li; Citrus Postharvest Green Mold: Recent Advances in Fungal Pathogenicity and Fruit Resistance. Microorganisms 2020, 8, 449, 10.3390/microorganisms8030449.
- Lucas, J.A.; Hawkins, N.J.; Fraaije, B.A. . The Evolution of Fungicide Resistance. In Advances in Applied Microbiology; Sariaslani, S., Gadd, G.M., Eds.; Academic Press: Cambridge, MA, USA, 2015; pp. Vol 90, pp. 29-92.
- Ren-Shui Liu; Hu Huang; Qiang Yang; Wang-Yi Liu; Purification of α-Sarcin and an Antifungal Protein from Mold (Aspergillus giganteus) by Chitin Affinity Chromatography. Protein Expression and Purification 2002, 25, 50-58, 10.1006/prep.2001.1608.
- T. Theis; M. Wedde; V. Meyer; U. Stahl; The Antifungal Protein from Aspergillus giganteus Causes Membrane Permeabilization. Antimicrobial Agents and Chemotherapy 2003, 47, 588-593, 10.1128/aac.47.2.588-593.2003.
- Henrietta Szappanos; Gyula Péter Szigeti; Balázs Pál; Zoltán Rusznák; Géza Szűcs; Éva Rajnavölgyi; József Balla; György Balla; Emőke Nagy; Éva Leiter; et al. The antifungal protein AFP secreted by Aspergillus giganteus does not cause detrimental effects on certain mammalian cells. Peptides 2006, 27, 1717-1725, 10.1016/j.peptides.2006.01.009.
- Ana Beatriz Moreno; Álvaro Martínez del Pozo; Marisé Borja; Blanca San Segundo; Activity of the Antifungal Protein from Aspergillus giganteus Against Botrytis cinerea. Phytopathology® 2003, 93, 1344-1353, 10.1094/phyto.2003.93.11.1344.
- Lucía Citores; Rosario Iglesias; Sara Ragucci; Antimo Di Maro; José M. Ferreras; Antifungal Activity of α-Sarcin against Penicillium digitatum: Proposal of a New Role for Fungal Ribotoxins. ACS Chemical Biology 2018, 13, 1978-1982, 10.1021/acschembio.8b00410.
- Feng Zhu; Yang-Kai Zhou; Zhao-Lin Ji; Xiao-Ren Chen; The Plant Ribosome-Inactivating Proteins Play Important Roles in Defense against Pathogens and Insect Pest Attacks. Frontiers in Plant Science 2018, 9, 146, 10.3389/fpls.2018.00146.
- Lucía Citores; Rosario Iglesias; José Ferreras; Antiviral Activity of Ribosome-Inactivating Proteins. Toxins 2021, 13, 80, 10.3390/toxins13020080.
- Lucía Citores; Rosario Iglesias; Carolina Gay; José Miguel Ferreras; Antifungal activity of the ribosome-inactivating protein BE27 from sugar beet (Beta vulgaris L.) against the green mouldPenicillium digitatum. Molecular Plant Pathology 2015, 17, 261-271, 10.1111/mpp.12278.
- Yongchun Li; Meirong Zhao; Zhi Zhang; Quantitative proteomics reveals the antifungal effect of canthin-6-one isolated from Ailanthus altissima against Fusarium oxysporum f. sp. cucumerinum in vitro. PLOS ONE 2021, 16, e0250712, 10.1371/journal.pone.0250712.
- Thomas Aumer; Sébastien N. Voisin; Thomas Knobloch; Céline Landon; Philippe Bulet; Impact of an Antifungal Insect Defensin on the Proteome of the Phytopathogenic Fungus Botrytis cinerea. Journal of Proteome Research 2020, 19, 1131-1146, 10.1021/acs.jproteome.9b00638.
- Liming Shi; Beibei Ge; Jinzi Wang; Binghua Liu; Jinjin Ma; Qiuhe Wei; Kecheng Zhang; iTRAQ-based proteomic analysis reveals the mechanisms of Botrytis cinerea controlled with Wuyiencin. BMC Microbiology 2019, 19, 1-14, 10.1186/s12866-019-1675-4.
- Mingyan Li; Chi Chen; Xiaoshuang Xia; Betchem Garba; Linlin Shang; Yun Wang; Proteomic analysis of the inhibitory effect of chitosan on Penicillium expansum. Food Science and Technology 2020, 40, 250-257, 10.1590/fst.40418.
- Isabel Martins; Adélia Varela; Luís M. T. Frija; Mónica A. S. Estevão; Sébastien Planchon; Jenny Renaut; Carlos A. M. Afonso; Cristina Silva Pereira; Proteomic Insights on the Metabolism of Penicillium janczewskii during the Biotransformation of the Plant Terpenoid Labdanolic Acid. Frontiers in Bioengineering and Biotechnology 2017, 5, 45, 10.3389/fbioe.2017.00045.
- Ting Zhou; Bishun Ye; Zhiqian Yan; Xiaohong Wang; Tongfei Lai; Uncovering proteomics changes of Penicillium expansum spores in response to decanal treatment by iTRAQ. Journal of Plant Pathology 2020, 102, 721-730, 10.1007/s42161-020-00486-6.
- Shu-Hua Lin; Pan Luo; En Yuan; Xiangdong Zhu; Bin Zhang; Xiaoyu Wu; Physiological and Proteomic Analysis of Penicillium digitatum in Response to X33 Antifungal Extract Treatment. Frontiers in Microbiology 2020, 11, 584331, 10.3389/fmicb.2020.584331.
- Yujun Zhou; Jianqiang Xu; Yuanye Zhu; Yabing Duan; Mingguo Zhou; Mechanism of Action of the Benzimidazole Fungicide on Fusarium graminearum: Interfering with Polymerization of Monomeric Tubulin But Not Polymerized Microtubule. Phytopathology 2016, 106, 807-813, 10.1094/phyto-08-15-0186-r.
- Di Pietro, A.; Roncero, M.I.G.; Roldán, M.C.R.. From Tools of Survival to Weapons of Destruction: The Role of Cell Wall-Degrading Enzymes in Plant Infection. In Plant Relationships. The Mycota (A Comprehensive Treatise on Fungi as Experimental Systems for Basic and Applied Research); Deising, H.B., Eds.; Springer: Berlin, Heidelberg, 2009; pp. Volume 5.
- Mario López-Pérez; Ana-Rosa Ballester; Luis González-Candelas; Identification and functional analysis ofPenicillium digitatumgenes putatively involved in virulence towards citrus fruit. Molecular Plant Pathology 2014, 16, 262-275, 10.1111/mpp.12179.
- Qiya Yang; Xin Qian; Solairaj Dhanasekaran; Nana Adwoa Serwah Boateng; Xueli Yan; Huimin Zhu; Fangtao He; Hongyin Zhang; Study on the Infection Mechanism of Penicillium Digitatum on Postharvest Citrus (Citrus Reticulata Blanco) Based on Transcriptomics. Microorganisms 2019, 7, 672, 10.3390/microorganisms7120672.
- Congyi Zhu; Yuying Wang; Xu Hu; Mengying Lei; Mingshuang Wang; Jiwu Zeng; Hongye-Ye Li; Zheyu Liu; Ting Zhou; Dongliang Yu; et al. Involvement of LaeA in the regulation of conidia production and stress responses in Penicillium digitatum. Journal of Basic Microbiology 2019, 60, 82-88, 10.1002/jobm.201900367.
- Mitchell Mutz; Terry Roemer; The GPI anchor pathway: a promising antifungal target?. Future Medicinal Chemistry 2016, 8, 1387-1391, 10.4155/fmc-2016-0110.
- Ruoxin Ruan; Mingshuang Wang; Xin Liu; Xuepeng Sun; Kuang-Ren Chung; Hongye Li; Functional analysis of two sterol regulatory element binding proteins in Penicillium digitatum. PLOS ONE 2017, 12, e0176485, 10.1371/journal.pone.0176485.
- Sara J. Blosser; Brittney Merriman; Nora Grahl; Dawoon Chung; Robert A. Cramer; Two C4-sterol methyl oxidases (Erg25) catalyse ergosterol intermediate demethylation and impact environmental stress adaptation in Aspergillus fumigatus. Microbiology 2014, 160, 2492-2506, 10.1099/mic.0.080440-0.
- Jonas Henrique Costa; Jaqueline Moraes Bazioli; João Guilherme De Moraes Pontes; Taícia Pacheco Fill; Penicillium digitatum infection mechanisms in citrus: What do we know so far?. Fungal Biology 2019, 123, 584-593, 10.1016/j.funbio.2019.05.004.
- D. Macarisin; L. Cohen; A. Eick; G. Rafael; E. Belausov; Michael Wisniewski; S. Droby; Penicillium digitatum Suppresses Production of Hydrogen Peroxide in Host Tissue During Infection of Citrus Fruit. Phytopathology® 2007, 97, 1491-1500, 10.1094/phyto-97-11-1491.
- Atiar Rahman; Suresh G. Kumar; Sang Woo Kim; Hye Jin Hwang; Yu Mi Baek; Sung Hak Lee; Hee Sun Hwang; Yun Hee Shon; Kyung Soo Nam; Jong Won Yun; et al. Proteomic analysis for inhibitory effect of chitosan oligosaccharides on 3T3-L1 adipocyte differentiation. PROTEOMICS 2008, 8, 569-581, 10.1002/pmic.200700888.
- Ioannis Stergiopoulos; Lute-Harm Zwiers; Maarten A. De Waard; Secretion of Natural and Synthetic Toxic Compounds from Filamentous Fungi by Membrane Transporters of the ATP-binding Cassette and Major Facilitator Superfamily. Netherlands Journal of Plant Pathology 2002, 108, 719-734, 10.1023/a:1020604716500.
- Paloma Sánchez-Torres; Molecular Mechanisms Underlying Fungicide Resistance in Citrus Postharvest Green Mold. Journal of Fungi 2021, 7, 783, 10.3390/jof7090783.
- Marta de Ramón-Carbonell; Paloma Sánchez-Torres; Penicillium digitatum MFS transporters can display different roles during pathogen-fruit interaction. International Journal of Food Microbiology 2020, 337, 108918, 10.1016/j.ijfoodmicro.2020.108918.
- Sandra Garrigues; Mónica Gandía; Attila Borics; Florentine Marx; Paloma Manzanares; Jose F. Marcos; Mapping and Identification of Antifungal Peptides in the Putative Antifungal Protein AfpB from the Filamentous Fungus Penicillium digitatum. Frontiers in Microbiology 2017, 8, 592, 10.3389/fmicb.2017.00592.
- Sandra Garrigues; Mónica Gandía; Crina Popa; Attila Borics; Florentine Marx; María Coca; Jose F. Marcos; Paloma Manzanares; Efficient production and characterization of the novel and highly active antifungal protein AfpB from Penicillium digitatum. Scientific Reports 2017, 7, 1-13, 10.1038/s41598-017-15277-w.
- Marina Marcet-Houben; Ana-Rosa Ballester; Beatriz de la Fuente; Eleonora Harries; Jose F Marcos; Luis González-Candelas; Toni Gabaldón; Genome sequence of the necrotrophic fungus Penicillium digitatum, the main postharvest pathogen of citrus. BMC Genomics 2012, 13, 646-646, 10.1186/1471-2164-13-646.
- Vera Meyer; Sascha Jung; Antifungal Peptides of the AFP Family Revisited: Are These Cannibal Toxins?. Microorganisms 2018, 6, 50, 10.3390/microorganisms6020050.
