Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Plant Sciences
Catalases (CATs) are considered the most potent (reactive oxygen species) ROS scavengers because of their strong affinity for H2O2.
- wheat
- CAT
- gene family
- genome-wide
- expression pattern
- abiotic stress
1. Introduction
Plant growth and development are affected by external abiotic and biotic stresses [1]. Environmental stresses alter cellular redox homeostasis, leading to the overproduction of reactive oxygen species (ROS), such as hydrogen peroxide (H2O2), hydroxyl radicals (OH−), superoxide (O2•), and other oxygen radicals [2][3]. ROS act as signaling molecules mediating many biological processes, but excessive accumulation of ROS in living plants can lead to oxidative stress damage to cells [4][5]. To adapt to the living environment, these highly toxic ROS must be converted into less reactive forms. Plants have evolved effective detoxification mechanisms, including non-enzymatic and enzymatic detoxification systems, to protect themselves from oxidative damage [2]. H2O2 is a major ROS and serves as an important messenger in abscisic acid (ABA) [6] and stress response pathways [7][8]. As the major mechanisms for removing ROS, the enzymatic systems include various types of antioxidant enzymes. Catalases (CATs) are considered the most potent ROS scavengers because of their strong affinity for H2O2 [3].
CATs have been found in almost all living organisms [9]; they have been studied extensively [3]. It have been illustrated that plant CAT gene expression is involved in the regulation of growth, development, and response to environmental stimuli [10][11][12]. In Arabidopsis, AtCAT genes encode a small family of proteins, including AtCAT1, AtCAT2and AtCAT3, which catalyze the decomposition of H2O2 and play an important role in controlling ROS homeostasis [13]. AtCAT1 expression is induced by ABA, which is mediated by MAPK cascades [14], but it does not seem to respond to circadian rhythms [15]. AtCAT2 is mainly expressed in leaves; it can be induced by light and cold, and may also be regulated by the circadian clock [15]. It has been reported that an AtCAT2 mutant (cat2), defective in AtCAT2 expression and, exhibiting 20% of the wild-type leaf catalase activity, accumulates more H2O2 than the wild type under normal growth conditions [16]. AtCAT3 is highly expressed in the whole plant at all developmental stages [17], is regulated by CPK8, and participates in ABA-mediated stomatal regulation in response to drought stress [6]. In rice (Oryza sativa), three CAT genes have been identified, including OsCATA, OsCATB, and OsCATC. Previous studies have demonstrated that OsCATA and OsCATC are the most stress-responsive members. The overexpression of OsCATA or OsCATC increases drought resistance in rice [8]. Moreover, OsCATC can be phosphorylated and activated by STRK1, improving both salt and oxidative tolerance in rice [10].
In addition, three CAT genes have been identified in tobacco (Nicotiana tabacum) [18]. Here, 3-AT (3-Amino-1,2,4-triazole) reduces catalase activity by affecting catalase mRNA abundance, whereas paraquat induces NtCAT3 expression. In sweet potato (Ipomoea batatas), SPCAT1 plays a role in H2O2 homeostasis in leaves and in the plant’s response to environmental stress [19]. Interestingly, ectopic expression of CAT can also affect CAT activity and affect plant resistance to adverse conditions. For example, the expression of a wheat catalase gene in rice can enhance tolerance to low temperatures [20]. In addition, the ectopic expression of maize CAT2 (ZmCAT2) in tobacco can induce CAT activity, improving pathogen resistance [21].
Several recent studies have firmly established that the ROS-scavenging enzymes can be mediated by several microRNAs (miRNAs) to improve oxidative stress tolerance in plants [22][23]. miRNAs are a class of endogenous non-coding RNAs (approximately 22 nucleotides) that participate in plant development and responses to environmental stress by negatively regulating target gene expression [24][25]. It was indicated that higher miR408 expression contributes to enhanced antioxidant capacity, and improve tolerance to salinity, cold, and oxidative stress in Arabidopsis [26]. Furthermore, miR166 has been found to be involved in Cd stress response through regulating its target gene OsHB4 in rice [27].
Common wheat is the most widely cultivated crop on Earth, contributing about a fifth of the total calories consumed by humans [28]. In a previous study, ten CAT genes have been systemically identified in wheat. TaCATs are classified into three classes, and their potential role in wheat development and different stress conditions have been revealed [20][29]. In addition, the increasing evidence that catalase and H2O2 coupled with the flour b* colour variation [30]. Studies have confirmed that TaCAT3-A1 is expressed throughout seed development and associated with flour b* color variation rather than TaCAT3-A2 [30][31]. In contrast to the research progress in other species, knowledge of the CAT genes in wheat is still limited [20]. The release of wheat whole-genome sequence data [28] enables systematic identification and analysis of wheat CAT genes at the genomic level.
2.Evolution, Expression Pattern and Function Analysis
Plant CAT genes usually comprise a small gene family [32]. As the genomes of many species have been sequenced, genome-wide analyses of CAT families have been widely carried out. There are three CAT genes in Arabidopsis [15], maize [33], rice [11], cucumber [32], two CAT genes in Hordeum vulgare [34], seven in cotton [35], and ten CAT genes in wheat [36]. Understanding the biological functions of TaCAT genes and the molecular mechanisms underlying their responses to stressful environments are helpful for developing new wheat cultivars with enhanced resistance to multiple environmental challenges. However, the CAT family in wheat has not been thoroughly researched because of the complexity of the wheat genome.
A comprehensive genomic analysis of wheat led to the identification of ten TaCAT genes; they were symbolized based on their positions on wheat chromosomes. Among the identified genes, nine were located on eight different chromosomes, while TaCAT3-U was provisionally mapped onto ChrUn (Figure 1). Gene tandem duplication and segmental duplication events are thought to be key mechanisms for increasing gene family diversity [29]. Gene tandem duplication events often occurred during plant evolution, leading to the expansion of gene families. If a chromosomal region within 200 kb contains two or more genes, the genes in this region are considered as the result of a tandem duplication event [30]. Interestingly, a tandem duplicated TaCAT gene pair (TaCAT3-A1, and TaCAT3-A2) was found in the wheat genome (Figure 1); these genes had no homologous copies in any other subgenome. In general, each wheat gene usually has three homologous copies in sub-genomes (A, B, and D) [37]. However, TaCAT1-A, TaCAT1-B, and TaCAT1-D were derived from 5A, 4B, and 4D, respectively. It was confirmed that this is because wheat A subgenomic progenitor had translocation that occurred between chromosomes 4AL and 5AL [29]. The results suggested that the TaCAT gene expansion was the result of genome polyploidization and gene duplications during evolution.
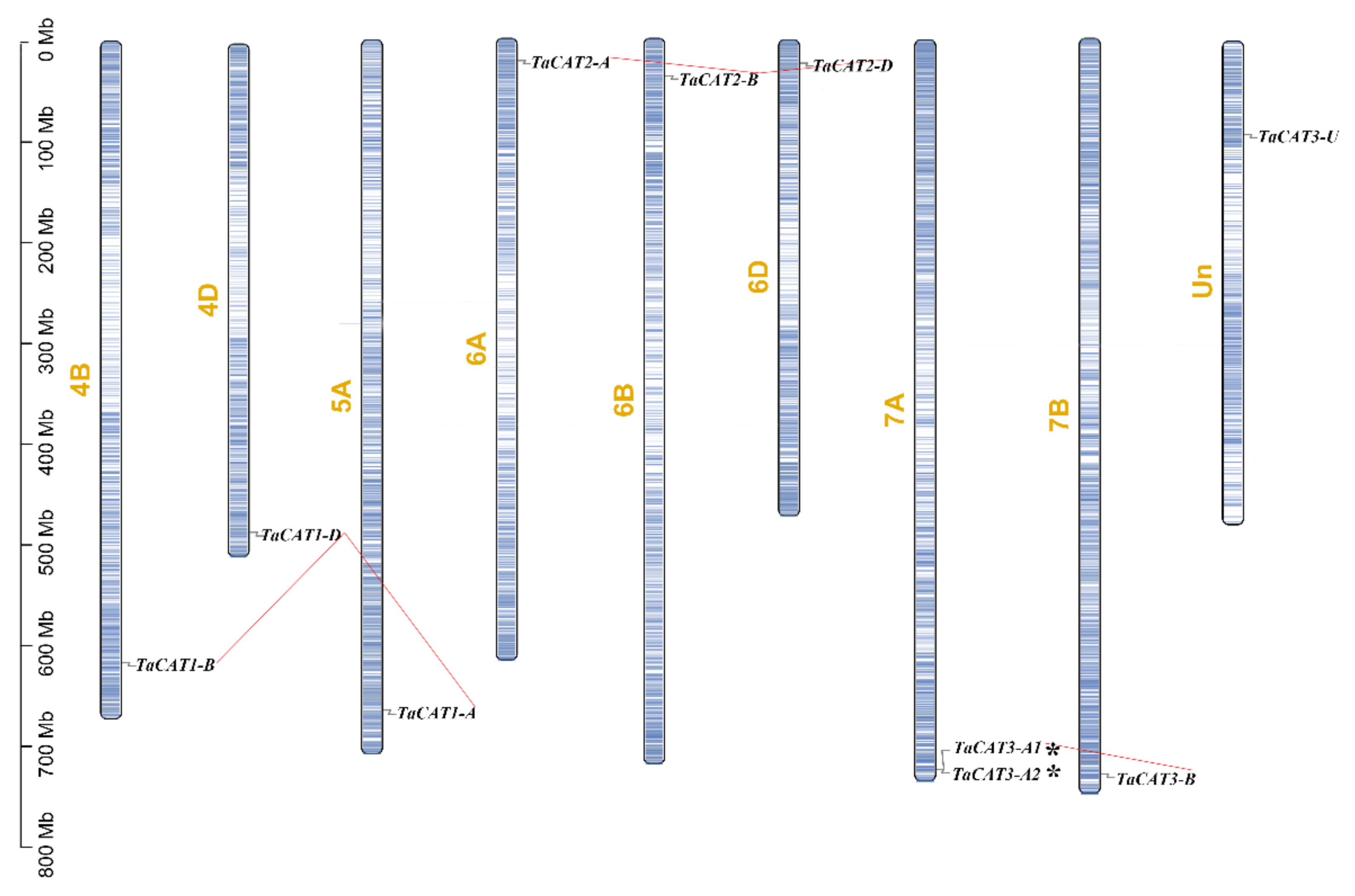
Figure 1. Schematic representations for chromosome distribution and duplication events of TaCAT genes. Chromosomal positions of Ten TaCAT genes were mapped on the basis of Wheat Genome (version IWGSC v1.0). Blue lines on each chromosome represented gene density. Segmental duplication events were indicated by red lines. * indicated the tandem duplication TaCAT gene pair. Only chromosomes that containing TaCAT genes were represented, and the chromosome number was indicated next to each chromosome.
Gene structure has been identified as one of the representative traces of gene family evolution [38]. The exons and introns of TaCAT genes were found to be different between monocots and dicots. All TaCAT genes possessed one to seven introns and two to seven exons (Figure 2b). In addition, a previous study suggested that one ancestral copy of a CAT gene had seven introns [39]. Together, these results confirmed that TaCAT genes had gone through exon and intron changes during their evolution. Additionally, the corresponding TaCAT proteins contained catalase and catalase-rel domains (Figure 2c), suggesting that these genes were conserved during evolution.
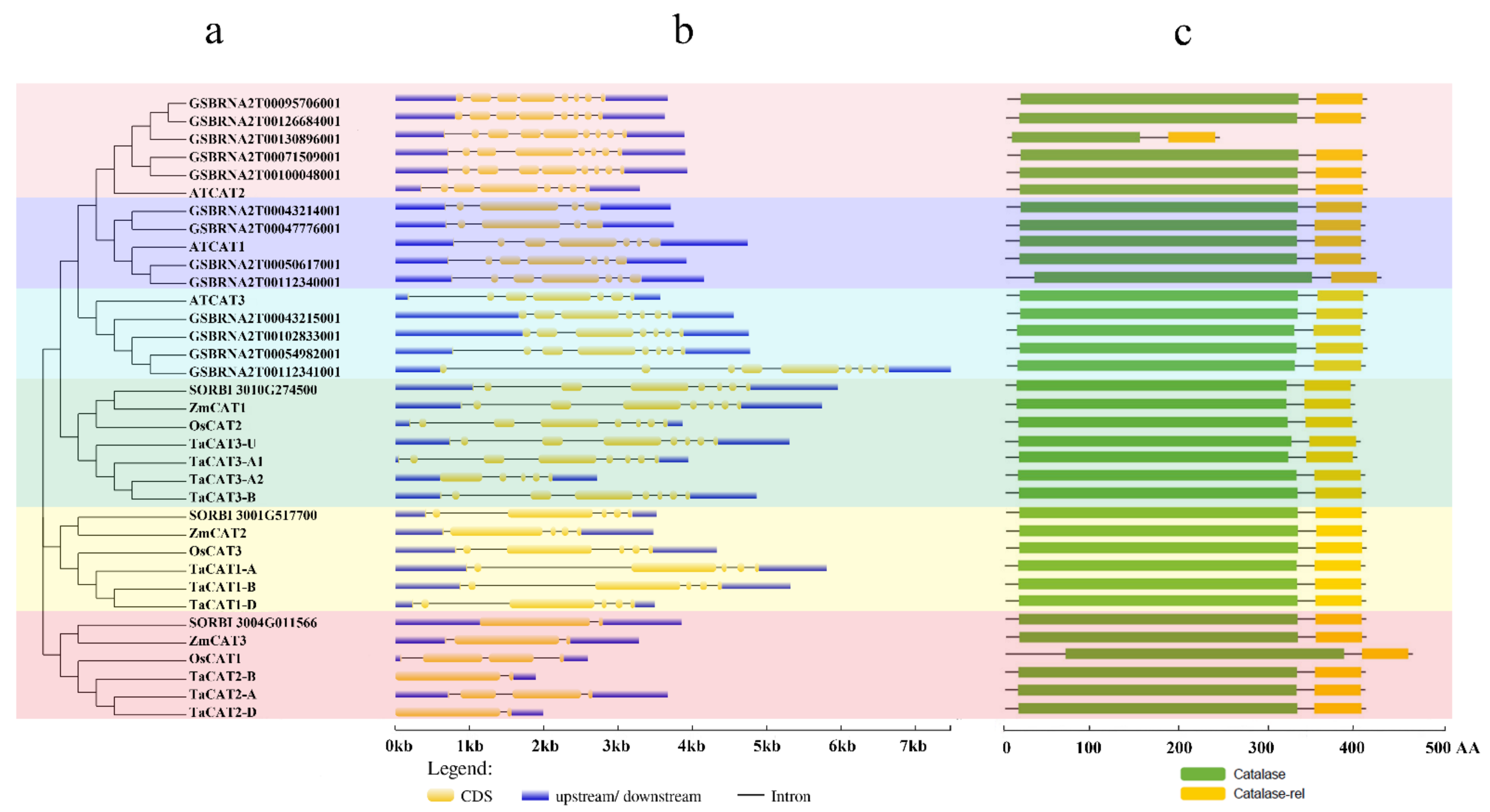
Figure 2. Phylogenetic relationships, gene structures, and conserved domains in CAT genes of Arabidopsis, B. napus, rice, sorghum, wheat, and maize. (a) Phylogenetic tree was constructed based on the full-length protein sequences of CAT genes using MEGA-X software with the calibration parameter of 1000; (b) exon–intron structure of CAT genes. The blue box represents the upstream/downstream sequences. The yellow boxes and the black horizontal lines represent the exons and introns, respectively; Scale indicates 1.0 kb; (c) schematic diagram of the conserved domain of CAT proteins. Catalase core domain (Catalase, PF00199) and catalase-related immune-responsive domain (Catalase-rel, PF06628) were shown in the blue and yellow boxes, respectively; Scale indicates 100 AA.
Gene expression is regulated by the complex interaction of many cis-acting elements and trans-acting factors that participate in various pathways [32]. Previous studies have confirmed that some CAT genes could be induced by different treatments such as cold [13][39], ABA [6][40][35], drought [6][13][36], and light [41]. However, little research has been carried out to investigate the cis-acting elements of TaCAT gene promoters. Furthermore, a promoter analysis of the TaCAT genes revealed the presence of various stress-responsive elements, such as ABRE, and cold responsive, defense and stress response, and anaerobic induction elements. Some TaCAT promoters also included an MYB-binding site (MBS), suggesting that some TaCATs might be regulated by the MYB transcription factor. In a previous study, it was observed that ROS mediated the control of plant stem cell fate and was key to stem cell maintenance and differentiation in Ababidopsis [42]. Moreover, some elements of meristem expression, cell cycle regulation, light response, SA, MeJA, and auxin responses were also found in some TaCAT gene promoters. This suggests that TaCAT genes might also be involved in plant growth and cell differentiation by regulating ROS metabolism networks.
ROS have been widely studied as important signaling molecules and toxic components in plants [3][4][43][44][45][46]. Thus, the potential roles of CAT genes encoding catalases that control the homeostasis of ROS are of great importance. Studies have suggested that CAT family members are involved in the regulation of plant growth, plant development, and environmental stress responses [3][11][32][47]. For example, heterologous expression of a TaCAT gene in rice improves tolerance against low temperatures [20]. It analyzed the transcript levels of TaCAT genes under heat, cold, NaCl, ABA, and mannitol treatments (Figure 3). The expression of all TaCAT genes changed dramatically when subjected to ABA treatment. These results indicated that TaCATs were closely related to the wheat ABA signaling pathway. Noteworthy, TaCAT1-A/B/D were found to be downregulated under NaCl and mannitol treatments, suggesting that they play specific roles when facing salt and drought stresses. It could not detect TaCAT3-A2 expression in any of the samples examined. Nevertheless, the functional roles of TaCAT genes require further investigation.

Figure 3. Expression analysis of TaCAT genes under different treatments by qRT-PCR. Relative expression levels of TaCATs in response to cold, heat, ABA, NaCl, and mannitol treatments for 0 h, 1 h, 2 h, 6 h, 12 h, and 24 h in the leaves at the three-leaf stage. Data were normalized with β-actin gene, and vertical bars indicate standard deviation error. Different letters indicate significant differences at p < 0.05 according to one-way ANOVA and post-hoc Tukey’s test.
In addition, valuable clues about the potential role of TaCAT genes in different tissues were obtained. The expression patterns of ten TaCAT genes were analyzed in different tissues at different developmental stages. The results demonstrated that TaCAT3-A1/B/U were constitutively expressed in almost all samples. TaCAT1-A/B/D showed a dramatically high level of expression in leaves, but it had a low expression level in the shoot axis at the milk grain stage (Figure 4). Furthermore, the expression of TaCAT2-A/B/D was high in leaves at the reproductive phase. In agreement with studies in other species [11][12][19][35][36][47], it was showed that some TaCAT genes were differentially expressed under various treatments, implying that TaCAT genes might be involved in environmental adaptation.
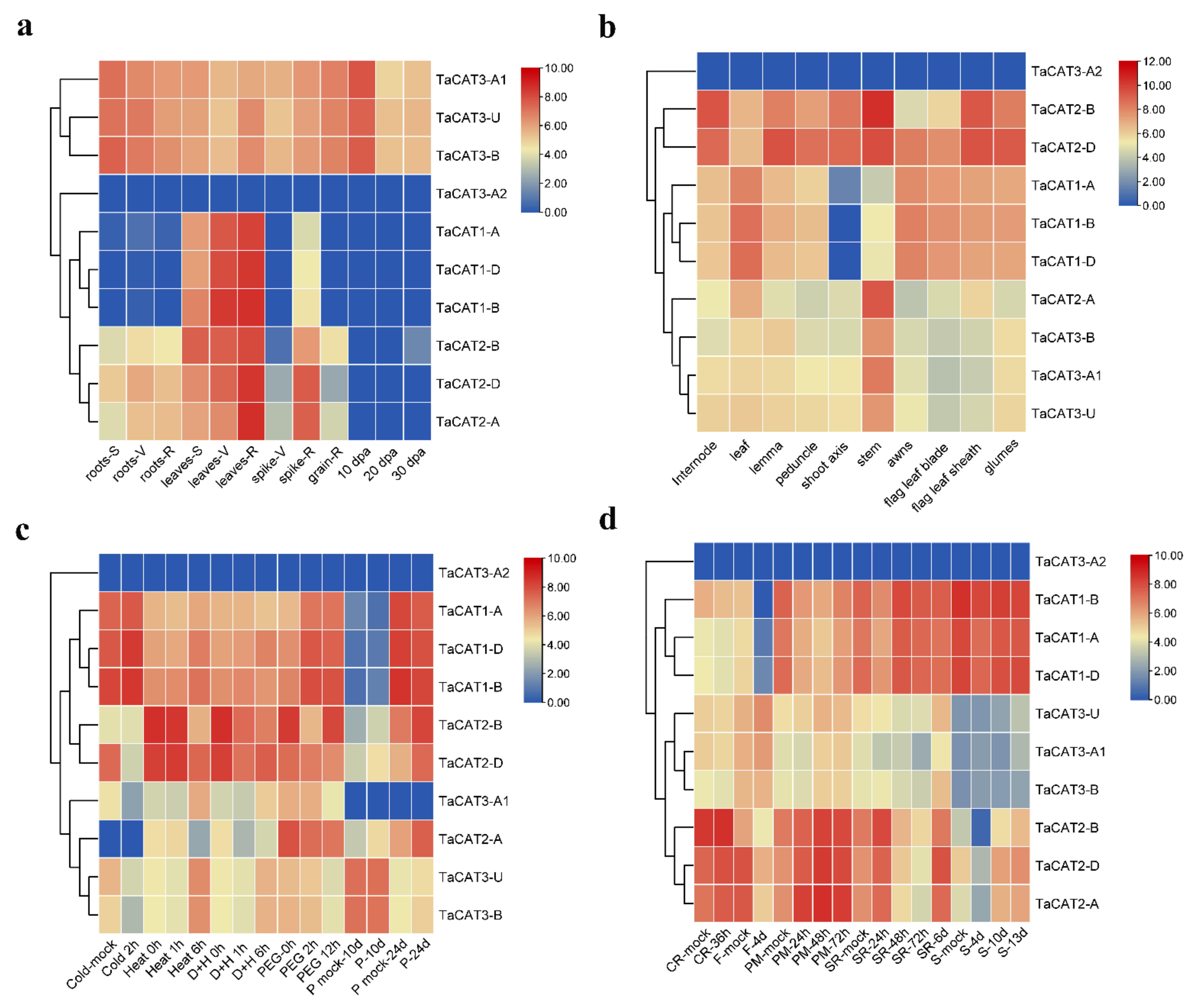
Figure 4. Expression analysis of TaCAT genes in different tissues of different developmental stages and stress treatments. (a) hierarchical clustering of expression profiles of wheat CAT genes in different tissues at seedling, vegetative, reproductive phase, and 10, 20, 30 dpa. S, seedling phase; V, vegetative phase; R, reproductive phase; dpa, day post-anthesis; (b) hierarchical clustering of expression profiles of TaCAT genes in different tissues at a milk grain stage; (c) hierarchical clustering of expression profiles of TaCAT genes under different abiotic stress treatments. D + H, drought and heat stress; P, phosphorus stress; (d) hierarchical clustering of expression profiles of TaCAT genes under different biotic stress treatments. CR, crown rot; F, fusarium hormones; PM, powdery mildew; SR, stripe rust; S, septoria.
Subcellular location is an important biological characteristic of proteins [48], and it is quite useful to understand the mechanisms underlying protein cellular activities. The catalases of Arabidopsis were found to be localized in the peroxisome (the main site of H2O2 production), whereas the rice catalases were predicted to be localized both in the cytoplasm and peroxisome [17]. Subcellular localization analyses showed that TaCAT2-A/B-GFP fusion proteins were localized in both the nucleus and cytoplasm, and that they formed protein aggregates in the cytoplasm (Figure 5). The formation of these aggregates and their molecular functions requires further study.
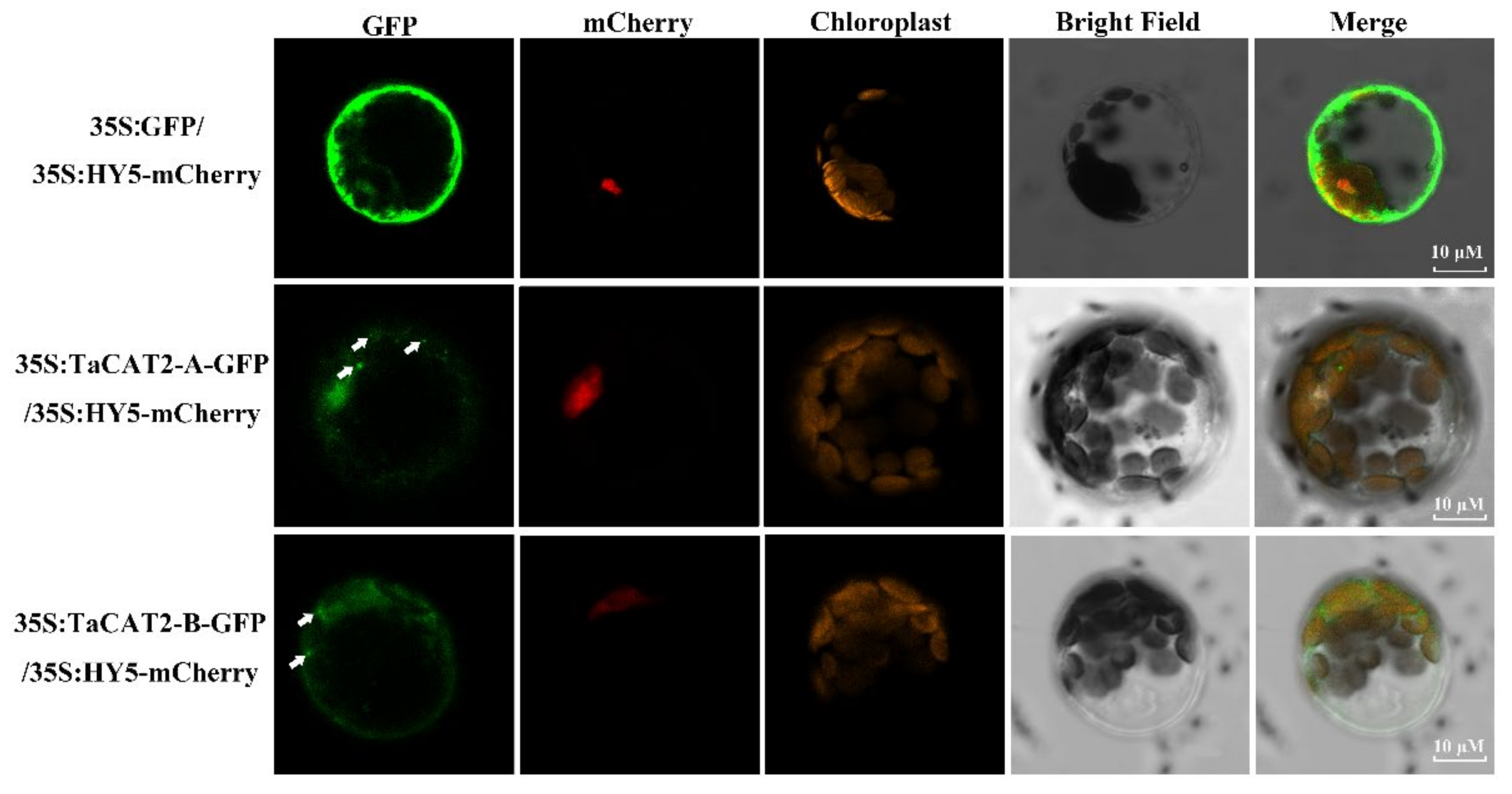
Figure 5. Subcellular localization analysis of TaCAT2-A and TaCAT2-B proteins in protoplast. TaCAT2-A-GFP and TaCAT2-B-GFP fusion protein were transiently expression driven by the 35S promoter in Arabidopsis protoplast. The fluorescence signals were observed under a laser scanning confocal microscopy. The green color represented the GFP fluorescence signals; the red and orange color represented the mCherry and the chloroplast fluorescence signals, respectively. The aggregated proteins were indicated by the white arrows. Scale bars were 10 μM.
miRNAs are single-stranded, noncoding RNAs that post-transcriptionally regulate target gene expression in plants and animals [49][50]. miRNAs participate in plant development and other physiological processes [51]. Previous studies have confirmed that CAT genes of rapeseed [36] and cotton [35] were targets of some miRNAs. An analysis of putative miRNA-targeting sites in the TaCAT genes showed that seven of these genes might be regulated by up to eight different miRNAs. Two members of the tae-miR395 family targeting three TaCATs (TaCAT1-A/B/D), while miR408 targeting two TaCATs (TaCAT2-A/B). miR395 has been reported to involve in cadmium detoxification in B. napus [52], resistance to leaf spot disease in apple [53], and upregulated under sulphate and phosphate (Pi) deficiency in Arabdopsis. miR408 has been reported to have pleiotropic effects on plant growth such as seed yield and leaf area in Arabdopsis [54], regulating heading time in wheat [55]. These studies suggest that these tae-miRNAs might play important roles against different stresses as well as participate in plant development by regulating the transcript level of the CAT genes in wheat.
This entry is adapted from the peer-reviewed paper 10.3390/ijms23010542
References
- Hirayama, T.; Shinozaki, K. Research on plant abiotic stress responses in the post-genome era: Past, present and future. Plant J. 2010, 61, 1041–1052.
- Malhan, D.; Bhatia, S.; Yadav, R.K. Genome wide gene expression analyses of Arabidopsis shoot stem cell niche cell populations. Plant Signal. Behav. 2015, 10.
- Mhamdi, A.; Queval, G.; Chaouch, S.; Vanderauwera, S.; Van Breusegem, F.; Noctor, G. Catalase function in plants: A focus on Arabidopsis mutants as stress-mimic models. J. Exp. Bot. 2010, 61, 4197–4220.
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399.
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498.
- Zou, J.-J.; Li, X.-D.; Ratnasekera, D.; Wang, C.; Liu, W.-X.; Song, L.-F.; Zhang, W.-Z.; Wu, W.-H. Arabidopsis CALCIUM-DEPENDENT PROTEIN KINASE8 and CATALASE3 Function in Abscisic Acid-Mediated Signaling and H2O2 Homeostasis in Stomatal Guard Cells under Drought Stress. Plant Cell 2015, 27, 1445–1460.
- Ma, L.; Zhang, H.; Sun, L.; Jiao, Y.; Zhang, G.; Miao, C.; Hao, F. NADPH oxidase AtrbohD and AtrbohF function in ROS-dependent regulation of Na(+)/K(+) homeostasis in Arabidopsis under salt stress. J. Exp. Bot. 2012, 63, 305–317.
- Schmidt, R.; Mieulet, D.; Hubberten, H.-M.; Obata, T.; Hoefgen, R.; Fernie, A.R.; Fisahn, J.; Segundo, B.S.; Guiderdoni, E.; Schippers, J.H.M.; et al. SALT-RESPONSIVE ERF1 Regulates Reactive Oxygen Species-Dependent Signaling during the Initial Response to Salt Stress in Rice. Plant Cell 2013, 25, 2115–2131.
- Frugoli, J.A.; Zhong, H.H.; Nuccio, M.L.; McCourt, P.; McPeek, M.A.; Thomas, T.L.; McClung, C.R. Catalase Is Encoded by a Multigene Family in Arabidopsis thaliana (L.) Heynh. Plant Physiol. 1996, 112, 327–336.
- Zhou, Y.-B.; Liu, C.; Tang, D.-Y.; Yan, L.; Wang, D.; Yang, Y.-Z.; Gui, J.-S.; Zhao, X.-Y.; Li, L.-G.; Tang, X.-D.; et al. The Receptor-Like Cytoplasmic Kinase STRK1 Phosphorylates and Activates CatC, Thereby Regulating H2O2 Homeostasis and Improving Salt Tolerance in Rice. Plant Cell 2018, 30, 1100–1118.
- Joo, J.; Lee, Y.H.; Song, S.I. Rice CatA, CatB, and CatC are involved in environmental stress response, root growth, and photorespiration, respectively. J. Plant Biol. 2014, 57, 375–382.
- Su, T.; Wang, P.P.; Li, H.J.; Zhao, Y.W.; Lu, Y.; Dai, P.; Ren, T.Q.; Wang, X.F.; Li, X.Z.; Shao, Q.; et al. The Arabidopsis catalase triple mutant reveals important roles of catalases and peroxisome-derived signaling in plant development. J. Integr. Plant Biol. 2018, 60, 591–607.
- Du, Y.-Y.; Wang, P.; Chen, J.; Song, C.-P. Comprehensive Functional Analysis of the Catalase Gene Family in Arabidopsis thaliana. J. Integr. Plant Biol. 2008, 50, 1318–1326.
- Xing, Y.; Jia, W.; Zhang, J. AtMKK1 mediates ABA-induced CAT1 expression and H2O2 production via AtMPK6-coupled signaling in Arabidopsis. Plant J. 2008, 54, 440–451.
- McClung, C. Regulation of Catalases in Arabidopsis. Free Radic. Biol. Med. 1997, 23, 489–496.
- Bueso, E.; Alejandro, S.; Carbonell, P.; Perez-Amador, M.A.; Fayos, J.; Belles, J.M.; Rodriguez, P.L.; Serrano, R. The lithium tolerance of the Arabidopsis cat2 mutant reveals a cross-talk between oxidative stress and ethylene. Plant J. 2007, 52, 1052–1065.
- Alam, N.B.; Ghosh, A. Comprehensive analysis and transcript profiling of Arabidopsis thaliana and Oryza sativa catalase gene family suggests their specific roles in development and stress responses. Plant Physiol. Biochem. 2018, 123, 54–64.
- Willekens, H.; Villarroel, R.; Van Montagu, M.; Inzé, D.; Van Camp, W. Molecular identification of catalases from Nicotianaplumbaginifolia (L.). FEBS Lett. 1994, 352, 79–83.
- Chen, H.-J.; Wu, S.-D.; Huang, G.-J.; Shen, C.-Y.; Afiyanti, M.; Li, W.-J.; Lin, Y.-H. Expression of a cloned sweet potato catalase SPCAT1 alleviates ethephon-mediated leaf senescence and H2O2 elevation. J. Plant Physiol. 2012, 169, 86–97.
- Matsumura, T.; Tabayashi, N.; Kamagata, Y.; Souma, C.; Saruyama, H. Wheat catalase expressed in transgenic rice can improve tolerance against low temperature stress. Physiol. Plant. 2002, 116, 317–327.
- Polidoros, A.; Mylona, P.; Scandalios, J. Transgenic tobacco plants expressing the maize Cat2 gene have altered catalase levels that affect plant-pathogen interactions and resistance to oxidative stress. Transgenic Res. 2001, 10, 555–569.
- Tang, J.; Chu, C. MicroRNAs in crop improvement: Fine-tuners for complex traits. Nat. Plants 2017, 3, 17077.
- He, J.; Jiang, Z.; Gao, L.; You, C.; Ma, X.; Wang, X.; Xu, X.; Mo, B.; Chen, X.; Liu, L. Genome-Wide Transcript and Small RNA Profiling Reveals Transcriptomic Responses to Heat Stress. Plant Physiol. 2019, 181, 609–629.
- Voinnet, O. Origin, Biogenesis, and Activity of Plant MicroRNAs. Cell 2009, 136, 669–687.
- Yu, Y.; Jia, T.; Chen, X. The ‘how’ and ‘where’ of plant microRNAs. New Phytol. 2017, 216, 1002–1017.
- Ma, C.; Burd, S.; Lers, A. miR408is involved in abiotic stress responses in Arabidopsis. Plant J. 2015, 84, 169–187.
- Ding, Y.F.; Gong, S.H.; Wang, Y.; Wang, F.J.; Bao, H.X.G.D.L.; Sun, J.W.; Cai, C.; Yi, K.K.; Chen, Z.X.; Zhu, C. MicroRNA166 Modulates Cadmium Tolerance and Accumulation in Rice. Plant Physiol. 2018, 177, 1691–1703.
- Appels, R.; Eversole, K.; Stein, N.; Feuillet, C.; Keller, B.; Rogers, J.; Khurana, J.P. Shifting the limits in wheat research and breeding using a fully annotated refrence genome. Science 2018, 361, 1–13.
- Tyagi, S.; Shumayla; Madhu; Singh, K.; Upadhyay, S.K. Molecular characterization revealed the role of catalases under abiotic and arsenic stress in bread wheat (Triticum aestivum L.). J. Hazard. Mater. 2021, 403, 123585.
- Crawford, A.C.; Francki, M.G. Chromosomal location of wheat genes of the carotenoid biosynthetic pathway and evidence for a catalase gene on chromosome 7A functionally associated with flour b* colour variation. Mol. Genet. Genom. 2013, 288, 483–493.
- Li, D.A.; Walker, E.; Francki, M.G. Identification of a member of the catalase multigene family on wheat chromosome 7A associated with flour b* colour and biological significance of allelic variation. Mol. Genet. Genom. 2015, 290, 2313–2324.
- Hu, L.; Yang, Y.; Jiang, L.; Liu, S. The catalase gene family in cucumber: Genome-wide identification and organization. Genet. Mol. Biol. 2016, 39, 408–415.
- Sytykiewicz, H. Transcriptional responses of catalase genes in maize seedlings exposed to cereal aphids’ herbivory. Biochem. Syst. Ecol. 2015, 60, 131–142.
- Kendall, A.C.; Keys, A.J.; Turner, J.C.; Lea, P.J.; Miflin, B.J. The isolation and characterisation of a catalase-deficient mutant of barley (Hordeum vulgare L.). Planta 1983, 159, 505–511.
- Wang, W.; Cheng, Y.; Chen, D.; Liu, D.; Hu, M.; Dong, J.; Zhang, X.; Song, L.; Shen, F. The Catalase Gene Family in Cotton: Genome-Wide Characterization and Bioinformatics Analysis. Cells 2019, 8, 86.
- Raza, A.; Su, W.; Gao, A.; Mehmood, S.; Hussain, M.; Nie, W.; Lv, Y.; Zou, X.; Zhang, X. Catalase (CAT) Gene Family in Rapeseed (Brassica napus L.): Genome-Wide Analysis, Identification, and Expression Pattern in Response to Multiple Hormones and Abiotic Stress Conditions. Int. J. Mol. Sci. 2021, 22, 4281.
- Feldman, M.; Levy, A. Allopolyploidy—A shaping force in the evolution of wheat genomes. Cytogenet. Genome Res. 2005, 109, 250–258.
- Hudson, K.A.; Hudson, M.E. A Classification of Basic Helix-Loop-Helix Transcription Factors of Soybean. Int. J. Genom. 2015, 2015, 603182-10.
- Frugoli, J.A.; McPeek, M.A.; Thomas, T.L.; McClung, C.R. Intron Loss and Gain During Evolution of the Catalase Gene Family in Angiosperms. Genet. 1998, 149, 355–365.
- Lam, B.C.H.; Blumwald, E. Domains as functional building blocks of plant proteins. Trends Plant Sci. 2002, 7, 544–549.
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49.
- Zhang, S.; Li, C.; Ren, H.; Zhao, T.; Li, Q.; Wang, S.; Zhang, Y.; Xiao, F.; Wang, X. BAK1 Mediates Light Intensity to Phosphorylate and Activate Catalases to Regulate Plant Growth and Development. Int. J. Mol. Sci. 2020, 21, 1437.
- Vighi, I.; Benitez, L.; Amaral, M.D.; Auler, P.; Moraes, G.; Rodrigues, G.; Da Maia, L.; Pinto, L.; Braga, E.J. Changes in gene expression and catalase activity in Oryza sativa L. under abiotic stress. Genet. Mol. Res. 2016, 15, 15.
- Schieber, M.; Chandel, N.S. ROS Function in Redox Signaling and Oxidative Stress. Curr. Biol. 2014, 24, R453–R462.
- Paunescu, R.A.; Bonciu, E.; Rosculete, E.; Paunescu, G.; Rosculete, C.A.; Babeanu, C. The Variability for the Biochemical Indicators at the Winter Wheat Assortment and Identifying the Sources with a High Antioxidant Activity. Plants 2021, 10, 2443.
- Elena, R.; Elena, B.; Aurelian, R.C.; Cătălin, D.Ș. Study on the yield and productivity elements of an assortment of winter wheat cultivated at ARDS Caracal. Ann. Univ. Craiova Agric. Montanology Cadastre Ser. 2020, 50, 192–197.
- Leung, D.W.M. Studies of Catalase in Plants Under Abiotic Stress. In Antioxidants and Antioxidant Enzymes in Higher Plants; Springer: Cham, Switzerland, 2018; pp. 27–39.
- Ang, L.H.; Chattopadhyay, S.; Wei, N.; Oyama, T.; Okada, K.; Batschauer, A.; Deng, X.W. Molecular Interaction between COP1 and HY5 Defines a Regulatory Switch for Light Control of Arabidopsis Development. Mol. Cell 1998, 1, 213–222.
- Ventura, A.; Young, A.G.; Winslow, M.M.; Lintault, L.; Meissner, A.; Erkeland, S.J.; Newman, J.; Bronson, R.T.; Crowley, D.; Stone, J.R.; et al. Targeted Deletion Reveals Essential and Overlapping Functions of the miR-17~92 Family of miRNA Clusters. Cell 2008, 132, 875–886.
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297.
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006, 57, 19–53.
- Zhang, L.W.; Song, J.B.; Shu, X.X.; Zhang, Y.; Yang, Z.M. miR395 is involved in detoxification of cadmium in Brassica napus. J. Hazard. Mater. 2013, 250-251, 204–211.
- Zhang, Q.; Li, Y.; Zhang, Y.; Wu, C.; Wang, S.; Hao, L.; Wang, S.; Li, T. Md-miR156ab and Md-miR395 Target WRKY Transcription Factors to Influence Apple Resistance to Leaf Spot Disease. Front. Plant Sci. 2017, 8, 526.
- Song, Z.; Zhang, L.; Wang, Y.; Li, H.; Li, S.; Zhao, H.; Zhang, H. Constitutive Expression of miR408 Improves Biomass and Seed Yield in Arabidopsis. Front. Plant Sci. 2018, 8, 2114.
- Zhao, X.Y.; Hong, P.; Wu, J.Y.; Chen, X.B.; Ye, X.G.; Pan, Y.Y.; Wang, J.; Zhang, X.S. The tae-miR408-Mediated Control of TaTOC1 Genes Transcription Is Required for the Regulation of Heading Time in Wheat. Plant Physiol. 2016, 170, 1578–1594.
This entry is offline, you can click here to edit this entry!
