Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
The MIP (molecularly imprinted polymer)-based biosensor can be considered an artificial antibody-integrated polymeric active layer that readily sustains stability in challenging testing chemical environments, such as high-temperature limits up to ~300 °C. Since general proteins are usually denatured in irreversible forms higher than ~80 °C, MIP-based biosensors are more stable in storage and even suitable for applications requiring a high-temperature range.
- molecularly imprinted polymer
- point-of-care test
- biomolecule
- oral disease
- wearable device
1. Introduction
Molecular diagnostics point-of-care (POC) is a technology belonging to the field of personalized healthcare and refers to clinical pathology tests for the diagnosis of disease [1]. Generally, it has been used to enhance the therapeutic effect by enabling an immediate test next to the patient, which was tested in the field of immunology and clinical chemistry [2]. POC devices are a type of in vitro diagnostic (IVD) medical device, designed for the purpose of diagnosing various diseases to determine prognosis, evaluating health status by medical treatment effect and even preventing disease [3]. The market growth for IVD devices can be attributed to the increasing proportion of the geriatric population and technological advancement in diagnostics [4]. Recently, during the COVID-19 pandemic, growing interest in public healthcare has rapidly boosted up rapid testing kits, anticipating the development of various types of devices with market expansion. Although the expensive cost of product development may defer research demand for POC testing (POCT), some progressive technologies are continuously introduced by merging existing portable sensing platforms in line with recently developed bioelectronic devices with suitable configurations in medical diagnosis applications [5,6,7]. One aspect of bioelectronics is the application of physicochemical signal transducers to detect substances at the molecular level and recognize interactions through signal processing. Biosensors used in POCT encompass a wide range of topics for the detection of analytes, various types of receptors, such as enzymes, antibodies, antigens, proteins at the interface of biological molecules and sensors. Therefore, since the POCT can be performed in close proximity to the location where the patient is being treated, emerging technologies as a potential alternative may replace the conventionally used laboratory-based diagnostic testing, including different combinations of components for sample handling and recognition elements. Thus far, the recent trend in the integration of diagnostic devices has moved to cost-effective programable tools for rapid and sensitive detection of biomarkers in biofluids, such as sweat, tear, saliva and urine [8,9,10]. However, many biomarkers in biological samples (i.e., biological fluids) are often present at very limited concentrations, coexisting with unwanted interfering species. Therefore, the detection of biomarkers usually requires highly qualified antibodies for sensitivity detection techniques together with sampling purification. To analyze one type of biomarker, enzyme-linked immunosorbent assay (ELISA) is a widely used immunological assay in diagnostic research [11], which provides quantitative data on specific proteins in serum samples. Despite its high specificity and low limit of detection (LOD), some drawbacks still arise from relatively long procedures with moderate reliability or expensive bioassay kits’ specified protocols, depending on the primarily designed binding affinity for different targets [12].
In this context, as a rational synthetic strategy and biomimetic design in the field of biotechnology, molecularly imprinted polymers (MIPs) have been revisited in response to the continuous demand for rapid, accurate and cost-effective analytical platforms. MIPs, crosslinked polymer matrices with molecular recognition sites formed by synthesizing in the presence of a target template, have received immense attention to guarantee affordable detection modules for target molecules for decades [13,14,15]. Historically, although viewed as an old material system, the MIP technology has progressed with a renewed field of research and expanded the area by combining emerging nanomaterials and advanced detection techniques with new applications [16,17,18]. Specifically, MIPs can be considered synthetic chemocavities or antibodies, as tailor-made artificial receptors that recognize and bind target molecules with high selectivity and chemical affinity [19,20,21]. The MIP matrices can be synthesized simply by polymerization of monomers forming a complex with target molecules, in which a relatively weak bonding was set between the template molecules and crosslinked monomers. In detail, starting with the prepolymer/template mixture, the spatial arrangement of MIPs can be determined by several well-known interactions, such as hydrogen bonds, Van der Waals forces, hydrophobic interactions and electrostatic interactions [22,23,24]. Subsequently, after the removal of template molecules from the crosslinked polymeric matrices, copious cavities with specified chemical end groups can be easily crafted, depending on complemental templates defined by size, shape and chemical functionality. Indeed, a large number of results on MIP techniques have reported newly developed molecular imprinting strategies with small molecules, such as sugars, steroids, pesticides, epitopes and amino acid derivatives [25,26,27]. These previous elaborated works have demonstrated the reliable capabilities of MIPs in highly targetable recognition systems on specific molecules, used in chemical sensors, analytic separations, solid-phase extractions, drug delivery systems, catalysts and library screening methods [28,29,30].
Having been progressively specialized in the field of biotechnology, MIPs were successfully commercialized for the solid phase of drugs and pesticides for an extraction toolbox to rebind small molecules toward improved sample refinement of chromatographic analysis [31,32]. In addition, molecular imprinting for other larger-scale substances in particular is also considered a candidate for expandible technology and remains under development with plenty of potential. Biomacromolecules, including antibodies, viruses, proteins, enzymes, nucleic acids and even living cells, can readily be imprinted in precisely designed polymer matrices with the help of other interfacial molecules or additives. However, the biomacromolecule approaches in the MIP system will confront serial problems with less reliability in the sophisticated capturing process because the classical bulk methodologies for target templates may fail to precisely recognize the protein target molecules. The lack of accessibility lies primarily in the intrinsic properties of protein molecules themselves. Complete rebinding may be difficult due to imprinted sites that differ from the original conformation by irreversible structural reconfiguration during polymerization [33]. In other words, templated proteins embedded in the polymerized matrix can be partially immobilized with strong physical bonds in the crosslinked polymeric network during the template removal step, which provides fewer rebinding sites [34]. Thus, improved techniques are needed, especially in protein imprinting processes, to prevent irreversible entrapment in 3D polymeric networks. Hence, the large number of uncontrolled interaction sites by the imprinted proteins may lead to cross-reactivity on the originally provided cavities along with non-specific adsorption. Biomacromolecule recognition systems have now become a new growing research area in MIP approaches, and the control of the biological environment associated with an appropriated function (i.e., artificial recognizable antibody) requires further development in practical use and biomedical diagnostic applications [35].
In view of the above, the MIP-based pseudo-immunoassays may be developed and may strengthen the biosensing capability and related POCT to measure the concentration of small and macromolecules with the help of specialized ‘artificial antibodies’ to antigen counterparts [36]. Thus, an enhanced accuracy of MIP-based POCT will accelerate the molecular diagnostic and has critical potential to play an important role in analytical tests in various fields with different macromolecular targets for more accurate results. For now, commonly used biosensors are mostly based on the detection of antigen–antibody interactions, which are evaluated and quantified with respect to each proposed transducing mechanism [37,38,39]. Moreover, most antibodies used are proteins, which are physically, chemically and biochemically unstable for use in a medical-grade immunoassay. Therefore, healthcare devices based on MIP-technology-based POCT may provide suitable access for specific patients by providing reliable results from artificial antibody-integrated biosensors (i.e., MIP-based testing). The MIP-based biosensing platform can be robust, sensitive and accurate to enable label-free detection of biomolecular analytes. Such beneficial embodiments will meet endless supply demands in the segmental market to develop MIP technology, leading to an integration of cost-effective portable POCT with an expansion of the overall industrial progression.
2. MIPs for Biomolecule Recognition: Concepts of POCT and Synthetic Approaches
2.1. Concepts of the MIP-Technology-Based Portable POCT Devices
The most important feature for MIP-enabled biosensors is the comparability that can be integrated with the existing systems, which provides high recognition ability [45]. The persistent durability allows MIPs to be used in various types of POCT applications, depending on the types of samples to be tested, as presented in Figure 1. Benchtop-scale POCT devices, incorporating MIP-based biosensors, are poised to transform the healthcare device platforms. Conventional biosensors combining biological elements have been produced in a form of chips, disposable strips, cartridges or electrodes in the application of POC devices [46,47]. However, in certain situations, the diagnostic devices have some limitations in new classes of POC devices [48]. For example, the short shelf life of biomolecular immobilized biosensors is less cost effective for manufactured products because they should be refrigerated in transport and storage [49]. Another potential issue might cause the low activity of biomolecular functions under harsh chemical conditions in some cases of biofluids or sampling, such as extreme changes in pH, saline or highly reactive solvents in certain treatment controls for the POCT devices [50], which critically affect the performance of biosensors by fundamental degradation of the disease-specific biomarkers [51]. Besides, although conventionally used bioreceptors are suitable for achieving selectivity, multiple processing with technical difficulties and delicate interface control are required in the implementation onto biosensors [52]. Since the immobilization of recognition sites is essential to transduce the signals for the operation of biosensors, advantageous materials and alternative strategies will be needed to resolve the shortcomings linked with conventional biomarker integration while achieving selectivity. By the aforementioned motivations, MIP techniques have been progressed for biosensor applications with carefully contrived design. The MIP-based biosensor can be considered an artificial antibody-integrated polymeric active layer that readily sustains stability in challenging testing chemical environments, such as high-temperature limits up to ~300 °C [53,54]. Since general proteins are usually denatured in irreversible forms higher than ~80 °C [55,56], MIP-based biosensors are more stable in storage and even suitable for applications requiring a high-temperature range. In the scene of biomolecule imprinting with low-cost materials, by taking advantage of the MIP technique to mimic biological sensing elements, such as antibodies and bioreceptors [57], a variety of single-target biosensors can be developed for diagnostic biosensors and assays for POCT devices [58]. Because the MIP-enabled biosensors extracted out the biological antibodies or other elements, the desired receptor surface can be tailored for relatively high selectivity and specificity. Compared to the biosensors integrated with natural antibodies, MIP-based biosensors have exhibited a comparable or even decreased limit of detection (LOD) with signal-to-noise enhancement and improved stability resulting in potential use for biosensing platforms [59]. The continued progress of MIP technology holds great potential with innovative key aspects of inexpensive, rapid and sensitive detection for desired POCT systems, providing other opportunities in demanding medical environments.
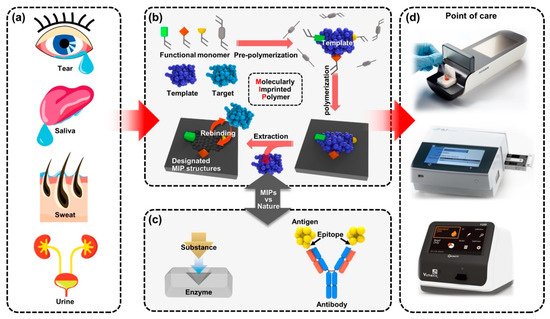
Figure 1. A new class of benchtop-scale POCT devices utilized by MIP-based biosensors for precision diagnostic technology to detect biomarkers in biofluids. (a) The four types of representative human biofluid reflecting health conditions; tears, saliva, sweat and urine. (b) Schematic illustration for fabricating the molecular imprinting system that contains the biorecognition sites and (c) the example of natural biorecognition system; enzyme–substrate complex (left) and antigen–antibody reaction (right); the biomimetic functional similarity of the MIP biosensing system is comparable to natural antibodies. (d) Various types of immunoassay-based benchtop-scale POCT devices.
2.2. Biomolecule Imprinted Polymers Based on Bulk Imprinting Techniques
At the beginning of the development of synthetic process for MIPs, target-oriented techniques, capable of recognizing and binding biomacromolecules (e.g., proteins), have been introduced along with practical use in numerous applications, including clinical diagnostics [60], drug delivery systems [61], proteomics and environmental analysis [62]. Despite the widespread research work on MIPs, there have been some limitations in the design of the MIP material system in protein detection by the intrinsic conditions of templated proteins, such as size, complexity and structural instability. Recently, however, the synthetic strategies for protein-based MIPs have been extensively developed to improve the selective recognition capability, named by bulk, suspension, emulsion and epitope imprinting, depending on the materials mainly featured [63,64,65]. As one synthetic process for protein-based MIPs, the so-called bulk imprinting polymerization is the most commonly accepted method with apparent advantage of the simplicity of the processing scheme. In designing bulk imprinting, by using the crosslinker and the functional monomer, protein molecules can be imprinted entirely on the growing polymer matrix with randomly distributed configuration, and subsequent extraction of the templates from the MIPs complete the process with high yield performance. Finally, for the collected MIP powders, the mechanical grinding process separates the bulk-imprinted polymers into the form of micron-scale particles or beads. This sequential process suggests a viable route to produce a large number of bulk MIP particles that can be used in several commercial applications [66]. However, for a typical bulk MIP system, a random free diffusion of the templates (i.e., small molecules) was subjected to the formation of microcavities in the densely networked MIP structures [67]. Therefore, bulk imprinting is adaptable for imprinting for small molecules because the adsorption/release of templated molecules is easily expected and represents relatively fast and reversible interactions, which add value to the small-molecule imprinted matrix as multiple-time reusable assay in cost-effective benefits [68]. On the other hand, a limited synthetic condition for the bulk-imprinted biomacromolecules has been reported due to partially trapped templated molecules in the polymer chains, commonly featured with a complex distribution in the mixed state prior to the polymerization. Such drawback lies in the limited diffusion rate of biomacromolecules from the nature of bulk MIP manufacturing system. Consequently, low diffusion rates attenuate the quality of MIPs with lacking accessibility on the rebinding sites. Moreover, the mechanical grinding process as a final stage is strictly controlled to maintain the original integrity of the prepared samples, that is, damages of the recognition sites reduce the adsorption capacity of the bulk MIP system. Although crushing films into smaller dimensional microparticles notably expands binding recognition sites, the irregular shapes and sizes of the resulting bulk MIP particles lead to less reliable signal detection for accurate biosensing of biomacromolecules with high precision [69]. Thus, to avoid the instability of protein-imprinted bulk MIPs, one key parameter can be the homogeneous combination between template sizes (i.e., the large size of proteins) of the incorporated monomers by careful design to provide protein recognition sites with high reproducibility. The nanoscale protein-imprinting polymer in uniform 3D bulk scale is inevitable for an improved binding site accessibility, meeting quality requirements for biosensor application on the appropriate analytical performance [70]. The advances of protein-imprinting technique have also expanded to direct construction of micro/nanoscale surface-imprinted MIP systems on planar surfaces with the development of combinatorial materials composition, which has suggested novel sensing mechanisms in various types of signal transducing systems [71]. In competition with the bulk MIP technique, other strategic templating processes have been introduced as alternative methods to resolve the problems with diffusion limitations, uniform features and improved selectivity.
2.3. Biomolecule-Imprinted Polymers Based on Surface Imprinting Techniques
As an effective way of integrating biomolecule-imprinted polymer into biosensor systems, newly developed surface imprinting techniques have been directly applied to transducing elements, such as chemically derived electric signals [72,73,74]. By the advantageous feature of the surface MIP system, the increased specific binding sites are exposed only on the surface of the polymer matrix for effective recognition, which thus accelerates mass transfer and accurate rebinding capacity (i.e., adsorption/desorption efficiency). To generate a protein-imprinted polymeric surface, a suitable monomer selection for the templates is a crucial factor in the rational design through high affinity of chemical composition for the advanced surface MIP system. Similar to the bulk MIP system, since the binding strength and stability between monomer and template depend on non-covalent weak forces, such as hydrogen/hydrophobic or electrostatic interactions, designing a template/monomer complex on the active surface area to effectively recognize the rebinding biomolecules is necessary. As shown in Figure 2, the interactions between proteins and monomers for constructing protein-imprinted polymers (i.e., artificial receptor formations) on the electrically conductive surface can be classified into several types: (i) formation of a prepolymerized complex on the electrode; (ii) sequential electropolymerization of functional monomers after template physisorption; and (iii) immobilization of the target protein using a complemental linker. As an easily accessible process, protein–monomer mixtures were introduced onto electrode surfaces using drop casting [75], spin casting [76] and spray coating [77]. After that, the prepolymerized complex was crosslinked under specific electrosynthesis conditions, and the templated protein could be extracted by physical or chemical methods to form steric cavities, constructing a surface-MIP recognition system (Figure 2a). On the other hand, Figure 2b represents another developed surface imprinting method that induces spontaneous adsorption of proteins to the electrode surface to increase templated cavities. The templates (i.e., proteins) built on the electrode surface can be imprinted by physicochemical interactions following the electrosynthetic process, in which the configured specific cavities are mostly on the surface of the MIP matrix. During this electropolymerization approach, the isoelectric point (pI) of the protein can be considered an important factor in designing sophisticated MIP–protein complexes. As an inherent property of proteins (i.e., amphiphilicity), the pI is usually defined by the pH value of a solution, at which the net charge is zero [78]. Thus, the electrostatic behavior of proteins with pH is subtle in the process because when the pH of a solution is higher than the pI value, the surface of the protein becomes predominantly negatively charged, resulting in a repulsive force on the same charged molecules. In contrast, in the case of lower pH of the solution than the pI value, the protein surface can be positively charged. However, under conditions with a pH value close to the pI, the attractive force prevails between the proteins by balancing the negative and positive charges, leading to the aggregation or precipitation of the protein [79,80]. To resolve this problem, the surface-MIP approach using a sacrificial carrier was demonstrated. By the fact that the aggregated form of proteins can lead to inadequate sensing properties in MIP-based biosensor systems, the formation of covalently immobilized proteins prior to electrodeposition of functional monomers on the electrode surface could obviously enhance the sensing performance for target proteins (Figure 2c). As one clear demonstration, the sequential molecular imprinting process of specific protein binding sites for selective recognition system in the surface-MIP structure is as follows [81]. First, the electrode surface could be chemically modified via a 4-ATP/DTSSP linker system and immobilized with a target protein (CDNF). After electrochemical polymerization with functional monomers, selective molecular cavities could be formed simply by the S–S bond cleavage process. This experimental approach implies that target protein immobilization can be facilitated by a simple combination of conventional chemistries (i.e., linkers such as self-assembled monolayers, SAMs) to create more uniform specific binding sites with finely tuned affinity [82,83,84], instead of random electropolymerization from the protein/monomer mixture. Notably, in this case, the optimized thickness of the surface-MIP film was precisely controlled during the electropolymerization process not to exceed the height of the immobilized target protein, which is the most important factor in avoiding irreversible entrapment of proteins in the imprinted polymer matrix [85].
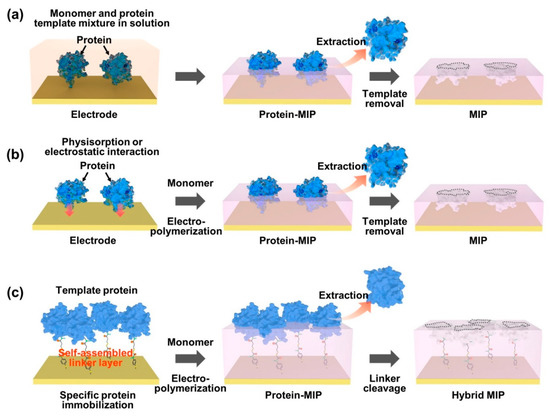
Figure 2. Representative strategies on the surface imprinting process to construct specific protein recognition cavities. In an appropriate design concept, the selective rebinding site can be generated by using a functional monomer for electropolymerization on a prepared electrode surface, which includes the formation of the pre-polymerization complex (a), the template physisorption (b) and the immobilization of the target protein (c).
2.4. Electrosynthetic Strategies for Biomolecule-Imprinted Polymers
As described in the previous section, direct electropolymerization has proved to be an efficient technique to construct surface-imprinted MIPs by the adequate combination of monomers and templates [86,87,88]. More importantly, the selection of biomolecular templates has not been limited within an allowed experimental condition and is ready to be applicable to state-of-the-art electropolymerization strategies on the electrically conductive electrode surfaces, such as RNAs, DNAs, peptides, aptamers, antibodies, proteins, viruses, bacteria and even living cells [89,90,91,92,93,94,95,96]. A suitable choice of monomers and templates plays a key role in the molecular design of electrodeposition techniques with delicate modulation of parameters for the MIP-enabled conducting polymer matrix. Thus, synergistic influences on surface MIPs have been evaluated as a result of highly specified analytical performance in the biosensing platforms [97,98]. As reported earlier, the main parameters for electrosynthetic process can be summarized as follows [99]: (i) voltage or current of applied potential; (ii) potential scan rate and periodic potential pulses during deposition cycles; and (iii) the restriction of electrical density on the electrode. Therefore, the electrosynthesis of conductive polymers in the surface-MIP system highly depends on the series of optimization by a control of the surface morphology, density and film thickness to tune the capability of charge transfer passing through the electrode [100]. Figure 3a represents a typical process of electropolymerization for protein-imprinted polymers. In the protein-imprinted polymerization step of the electrosynthesis process, the stacking monomer layers and boundaries define the shape of the complemental recognition sites according to the size of biomolecules, such as proteins with embedded functional groups. Thus, sequentially designed processing steps with tailored compositions can be important to define the exposed areas of the cavities and controlled distributions of biomolecules prior to the electrodeposition of monomers. Such electroactive monomers on a prepared electrode surface to be grown as an electrically conductive polymeric matrix should be carefully selected according to the electrosynthesis conditions because there are many combinatorial options with other binding assistant chemicals, including phenol, o-aminophenol, o-phenylenediamine, aminophenyl boronic acid, scopoletin, aniline, pyrrole, 3,4-ethylene dioxythiophene, 2,2′-bithiophene-5-carboxylic acid and dopamine, as previously reported [101,102,103,104]. For example, as illustrated in Figure 3b, a variety of combinations has been demonstrated by using o-phenylenediamine (o-PD) on template proteins for the surface-MIP integration [105,106]. Moreover, based on a computational approach, Raziq et al. recently demonstrated that o-PD could be a reasonable choice for biologically active MIPs compared to a set of molecules, such 3,4-ethylenedioxythiophen (EDOT) and dopamine (DA), in binding to the SARS-CoV-2 viral protein [107]. Looking into the detailed performance, the Glide empirical scoring function (GScore) values for the scoring binding pose of other monomers (i.e., o-PD, dopamine and EDOT) docked to SARS-CoV-2 nucleoprotein (ncovNP) were similar, in the ranges of −25.2 and −29.5 kJ mol−1, confirming that they could form stable pre-polymerized complexes with ncovNP. As a result of the quantum chemical calculation, the H-bond interactions on the ncovNP molecules adjunct with NH2 groups were determined decisively; the o-PD monomer was found to be a superior option compared to other monomers. This computational modeling approach can be highly useful and expanded to the advanced designing of MIPs, especially for the electrosynthetic process, because the molecular reaction and energy startup guidance based on the predominant parametric assumptions might be derived without repetitive control experiments [108]. In the biomolecular imprinting field, this high-end computational approach may be advantageous in rapid development in choosing correct parameters between the template and functional monomer to realize MIP-based biosensors, yielding highly selective recognition sites by scrutinizing the critical interaction energies [109]. By doing this, the electrosynthetic strategies for viral-protein-based MIPs (i.e., SARS-CoV-2) can contribute to producing a new concept of POCT devices to fully utilize the electrically operational transducers [110], which is under development with the popularly introduced small-scale device integrated with microchips for the wearable or skin-attachable format [111]. We will discuss this in more detail in the following sections.
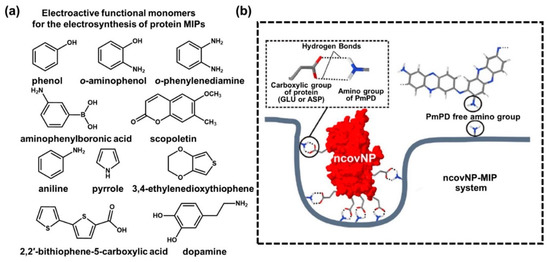
Figure 3. (a) Representative electroactive monomers used for implementation of the MIP-based biorecognition system; chemical structures of each functional monomer are listed as follows: phenol, o-aminophenol, o-phenylenediamine, aminophenylboronic acid, scopoletin, aniline, pyrrole, 3,4-ethylenedioxythiophene, 2,2′-bithiophene-5-carboxylic acid and dopamine. (b) Schematic illustration of the ncovNP–MIP system to form multiple non-covalent bonds between ncovNP and cavity. Reproduced with permission from [80]. Copyright Elsevier, 2020.
This entry is adapted from the peer-reviewed paper 10.3390/bios12030136
This entry is offline, you can click here to edit this entry!
