Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Biochemistry & Molecular Biology
A GH10 β-1,4-endoxylanase (XynSOS), from the ascomycetous fungus Talaromyces amestolkiae, has been heterologously produced in Pichia pastoris, purified, and characterized. rXynSOS is a highly glycosylated monomeric enzyme of 53 kDa that contains a functional CBM1 domain and shows its optimal activity on azurine cross-linked (AZCL)–beechwood xylan at 70 °C and pH 5. rXynSOS was capable of transglycosylating phenolic compounds, although with low efficiencies.
- antioxidants
- fungal enzyme
- glycosylation
1. Cloning, Production, and Purification of rXynSOS
A putative enzyme from the GH10 family, called g9427 and re-named as β-1,4-endoxylanase XynSOS, was detected in very low amounts when T. amestolkiae was grown in Mandels medium with microcrystalline cellulose, slurry, and xylan as carbon sources [25]. The xynSOS gene was located in the genome of the fungus [25] comprising 1445 bp, 3 introns, and a signal peptide region coding for 19 amino acids. The mature XynSOS protein is composed of 389 amino acids and contains a catalytic domain that shows high identity with other GH10 enzymes (HMMER dbCAN2, E-value 3.6 × 10−98) and a C-terminal Carbohydrate-Binding Module (CBM) assigned to family 1 (HMMER dbCAN2, E-value 8.4 × 10−17) and connected by a Ser/Thr rich linker region, as previously described in other GH10 endoxylanases [30].
The xynSOS gene without the three introns and lacking the signal peptide was expressed in P. pastoris GS115, since several GHs from this fungus have been successfully expressed in this model yeast to increase their production and facilitate their purification [26]. The clone with the highest recombinant XynSOS (rXynSOS) expression was selected for production, reaching a maximal endoxylanase activity against AZCL–beechwood xylan of 180 AU/min in the 9th day of culture in YEPS liquid medium (Figure 1a). The protein was purified from the crude extracts of P. pastoris in one single step using anion-exchange chromatography (Figure 1b), obtaining high-purification yields (recovered activity: 72%).
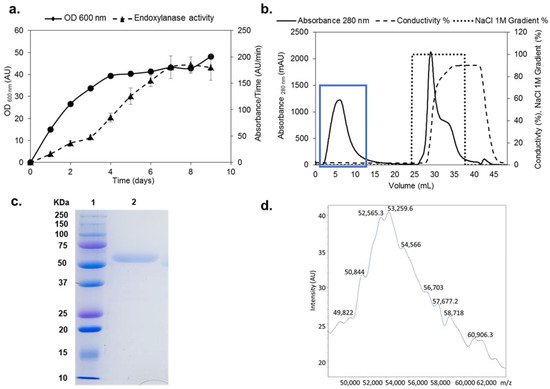
Figure 1. Production, purification, and characterization of rXynSOS. (a) Endoxylanase activity against AZCL–beechwood xylan and OD600 nm of the selected rXynSOS P. pastoris clone grown for 9 days in YEPS medium with methanol induction. (b) Purification of rXynSOS from P. pastoris crude extracts by anion-exchange chromatography. Protein absorbance through the NaCl gradient was monitored. The enzyme was purified in one single step. The blue rectangle indicates the peak of enzyme elution. (c) Determination of rXynSOS estimated molecular mass (Mw) by SDS-PAGE with the molecular weight marker displayed in lane 1. (d) MALDI-TOF spectrum of rXynSOS showing the profile of a glycosylated protein.
2. Physicochemical Properties of rXynSOS
The molecular mass (Mw) of rXynSOS determined by SDS-PAGE (Figure 1c) was around 50 kDa. This Mw is higher than the calculated by the ProtParam server from the rXynSOS amino acid sequence (41.57 kDa), which is probably due to the higher glycosylation levels of P. pastoris proteins [31]. The accurate Mw of the enzyme determined by MALDI-TOF was around 53 kDa (Figure 1d), showing a wide signal with multiple poorly defined peaks. This heterogeneous pattern is due to glycosylation, since 32 sites of O-glycosylation and one site of N-glycosylation were predicted in rXynSOS from its amino acid sequence. The Mw calculated by size exclusion chromatography was 55 kDa, indicating that rXynSOS is a monomeric protein in solution.
The maximum endoxylanase activity against AZCL-beechwood as substrate was observed at pH 5 and 70 °C. These values are comparable to those described for other fungal GH10 endoxylanases, ranging between 60 and 80 °C for the optimal temperature and pH 4.5 and 6 for the optimal pH [32,33,34,35,36,37].
The analysis of the functionality of the predicted rXynSOS CBM1 domain carried out after up to 2 h incubation of the enzyme with microcrystalline cellulose showed that the endoxylanase activity in the supernatant decreased very quickly, remaining only 27% after 10 min and 15% after 1 h, respectively. This result indicates that this CBM1 domain is functional and allows the enzyme to strongly bind to cellulose. Previous studies have postulated that the CBM1 domain enables GH10 endoxylanases to approach lignocellulosic biomass and bind to its crystalline cellulose, facilitating the degradation of the surrounding xylan [30,38].
3. Substrate Specificity and Kinetics of rXynSOS
The potential of rXynSOS to break down a broad range of substrates is displayed in Table 1. The enzyme was highly active against beechwood xylan (132.33 U mg−1) and especially against wheat arabinoxylan (149.15 U mg−1). This small preference for branched over linear xylans has also been reported for the GH10 endoxylanase XynD from Penicillium funiculosum [34]. rXynSOS was also able to hydrolyze CMC, although much less efficiently than the xylan substrates, as it was described with the GH10 endoxylanase AFUMN-GH10 from Aspergillus fumigatus [32]. Nevertheless, the enzyme could not hydrolyze microcrystalline cellulose and cellobiose, which are other specific substrates for enzymes with cellulolytic activity. The analysis on p-nitrophenyl-derived sugars revealed that rXynSOS can break down all four that were tested, being highly active on pNPX2 but showing lower hydrolytic activity against pNPX, pNPG2, and pNPG. As expected, a tendency to hydrolyze longer substrates was observed, since endoxylanase activity was better with xylans, and it was also higher on pNPX2 and pNPG2 than on pNPX and pNPG, respectively.
Table 1. Endoxylanase-specific activities of rXynSOS against different substrates.
| Substrate | Specific Activity (U mg−1) |
|---|---|
| Beechwood xylan (0.8%) | 132.33 ± 11.6 |
| Wheat arabinoxylan (0.8%) | 149.15 ± 3.45 |
| CMC (0.8%) | 1.25 ± 0.35 |
| Microcrystalline cellulose (0.8%) | Not active |
| Cellobiose (0.1%) | Not active |
| pNPX2 (0.1%) | 56.97 ± 3.96 |
| pNPX (0.1%) | 1.81 ± 0.06 |
| pNPG2 (0.1%) | 0.82 ± 0.01 |
| pNPG (0.1%) | 0.03 ± 0.001 |
Overall, these results confirm rXynSOS substrate versatility, which is in agreement with the capabilities of GH10 endoxylanases to hydrolyze β-1,4 linkages between xylopyranose residues located in the surroundings of side chains and even some cellulosic substrates [4,8,10]. In contrast, GH11 endoxylanases are usually very specific, as is the case of the endoxylanase XynM from the same T. amestolkiae strain, which was able to degrade beechwood xylan but no other substrates such as pNPX, pNPG, and CMC [12].
The kinetic parameters calculated for the best rXynSOS substrates, as well as for CMC as a representative of cellulose activity, are displayed in Table 2. The enzyme showed a high affinity for beechwood xylan and wheat arabinoxylan, with Km values of 1.07 and 1.94 g L−1, respectively, which are better than the ones reported for most fungal GH10 endoxylanases [32,34,36,37]. The catalytic efficiencies (kcat/Km) of rXynSOS on beechwood xylan (134.69 s−1 g−1 L) and wheat arabinoxylan (84.65 s−1 g−1 L) are in the same range as those of the GH10 endoxylanases Xyn10A from Penicillium oxalicum [36], PspXyn10 from Penicillum sp. [37], and XynD from P. funiculosum [34]. However, GH10 endoxylanases Xyn10B from P. oxalicum [36] and AFUMN-GH10 from A. fumigatus [32] exhibit unusually higher activities on beechwood xylan. In addition, the highest affinity (0.12 mM, 0.05 g L−1) and catalytic efficiency (880.04 s−1 mM−1, 2181.45 s−1 g−1 L) of rXynSOS were obtained with pNPX2 as substrate. Regarding the kinetic difference of GH10 and GH11 endoxylanases from T. amestolkiae, the results indicate that rXynSOS (GH10) is much more efficient than XynM (GH11) in the hydrolysis of beechwood xylan [12], in which the kcat/Km values are 134.69 and 7.76 s−1 g−1 L, respectively.
Table 2. Kinetic parameters of rXynSOS with different substrates.
| Substrate | Km (g L−1) | kcat (s−1) | kcat/Km (s−1 g−1 L) |
|---|---|---|---|
| Beechwood xylan | 1.07 | 144.25 | 134.69 |
| Wheat arabinoxylan | 1.94 | 163.93 | 84.65 |
| CMC | 51.29 | 7.90 | 0.15 |
| pNPX2 | 0.05 g L−1 0.12 mM |
102.96 | 2181.45 s−1 g−1 L 880.04 mM−1 |
4. Transglycosylation Potential of rXynSOS
The transglycosylation capacity of rXynSOS was assayed using pNPX2 as a xylobiose donor and vanillyl alcohol, 2-hydroxybenzyl alcohol, hydroquinone, and gallic acid as acceptors chosen on the basis of their interesting antioxidant properties and other health benefits [39,40]. The results showed that rXynSOS was able to transglycosylate all the acceptors assayed, but with low efficiencies, since the formation of glycosides was barely detectable by TLC and only confirmed by ESI-MS. Considering the wide substrate specificity observed in the enzyme’s hydrolytic activity, pNPX, pNPG, and pNPG2 were tested as donors of xylose, glucose, and cellobiose, respectively, for the transglycosylation of vanillyl alcohol. Despite the low efficiencies, ESI-MS spectra disclosed the production of glycosides for all the selected donors.
Although rXynSOS can transglycosylate different phenolic compounds employing a broad spectrum of sugar donors, its efficiency needs to be increased to enable its biotechnological application. The optimization of reaction conditions toward an enhanced transglycosylation/hydrolysis ratio is one of the strategies to improve glycoside yields [41]; however, the glycosynthase strategy based on eliminating the hydrolytic activity of the enzymes through protein engineering has been proven to be a better approach [20,41]. Therefore, a novel glycosynthase derived from rXynSOS was designed.
5. Conversion of rXynSOS into Its Glycosynthase Variants
Two glycosynthase variants of rXynSOS were developed by replacing its nucleophile catalytic residue by an inert one. The identification of rXynSOS nucleophile residue, a glutamic acid at position 236, was carried out via Clustal Omega alignment of XynSOS amino acid sequence with the sequences of three well-characterized fungal GH10 endoxylanases. Then, this residue was substituted by directed mutagenesis by glycine (rXynSOS-E236G) and serine (rXynSOS-E236S), since these mutations have been reported to generally produce higher glycosylation yields [18,42].
The two novel rXynSOS glycosynthase variants, which need activated glycosyl donors such as glycosyl fluorides to act on and catalyze the glycosylation reactions [19], were produced in P. pastoris GS115, selecting the clones by their higher total extracellular protein concentrations, since they no longer have hydrolytic activity (Figure 2a). The glycosynthase variants were purified in one single step using anion-exchange chromatography, following the same protocol as for the native rXynSOS, and their purity was confirmed by SDS-PAGE (Figure 2b).
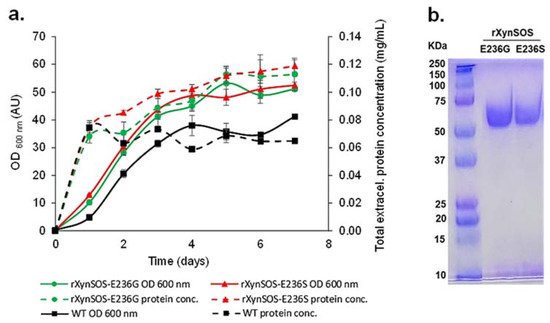
Figure 2. Production and purification of rXynSOS-E236G and rXynSOS-E236S glycosynthase variants. (a) Total extracellular protein concentration and OD600 nm of wild-type (WT) P. pastoris and the selected rXynSOS-E236G and rXynSOS-E236S-producing clones. Cultures were grown for 7 days in YEPS medium with methanol induction. (b) SDS-PAGE analysis of purified rXynSOS-E236G (lane 2) and rXynSOS-E236S (lane 3) variants. Molecular weight marker is displayed in lane 1.
The glycosylation activity of rXynSOS glycosynthase variants was studied in reactions with X2F as donor and pNPX2 as acceptor. Most of the glycosynthases that have been described are able to use pNP sugars as good acceptors for glycosylation, giving rise to pNP-oligosaccharides of different lengths [18]. Surprisingly, pNPX2 was not a good acceptor molecule for neither of the glycosynthase variants. In the case of rXynSOS-E236S, no products were detected either by TLC or ESI-MS, while for rXynSOS-E236G, the synthesis of pNPX4 and pNPX6 could only be confirmed by ESI-MS but not by TLC. To generate more conclusive data, we performed the reactions with vanillyl alcohol as an alternative acceptor molecule. The TLC results showed the expected glycosylated product for rXynSOS-E236G, which was also confirmed by ESI-MS. Nevertheless, rXynSOS-E236S was, again, unable to glycosylate the acceptor. The same behavior has been observed with other glycosynthases, being the synthetic activity of the serine mutants very low compared to the one of glycine variants [43]. This could be explained considering previous studies, which indicated that the rigid serine side chain might be an obstacle in the departure of the fluorine coming from the glycosyl fluorides. Moreover, the lack of side chain in glycine could also lead to a reduced steric hindrance, hosting the reaction more efficiently [18]. The detail of the catalytic nucleophile residue in the three-dimensional model of rXynSOS supports this hypothesis (Figure 3). Based on these results, the rXynSOS-E236G glycosynthase variant was selected for the following experiments.
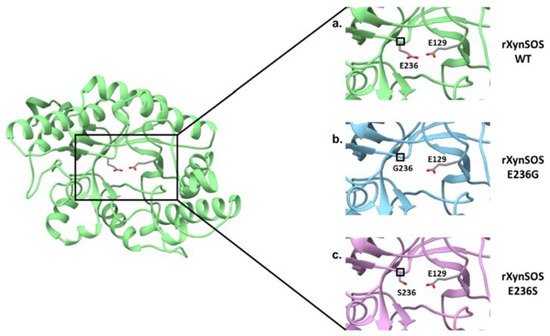
Figure 3. Conversion of T. amestolkiae rXynSOS into the glycosynthase variants rXynSOS-E236G and rXynSOSE236S. The three-dimensional model of the rXynSOS GH10 domain was obtained with SWISS-MODEL. The server selected as a template the PDB structure 6Q8M (GH10 endoxylanase from Aspergillus aculeatus), which has a sequence identity of 65.35% and resulted in a model of QMEAN 0.86. The catalytic amino acids E236 and E129 are displayed (a). Substitutions of the nucleophile catalytic residue E236 for G236 (lacking the side chain, b) and for S236 (shorter side chain, c) are shown to illustrate their effect in eliminating or reducing the hydrolytic capacity of the enzyme.
6. Oligosaccharides Synthesis by the Glycosynthase Variant rXynSOS-E236G
Xylooligosaccharides (XOS) have been described as emerging prebiotics that have important health benefits, such as immunomodulatory, antitumoral, and antimicrobial activities [44]. XOS are obtained by chemical or enzymatic methods, being the enzymatic approach the preferred one [18,45]. In the rXynSOS-E236G reaction using pNPX2 as an acceptor and X2F as a donor described before, several peaks of mass corresponding to XOS were detected by ESI-MS (Figure S8a). Taking into account this finding, we hypothesized that the synthesis of XOS was favored over the formation of pNP-XOS. To further study this, we conducted a reaction with X2F as the only reactant molecule (no acceptor was added), in which the synthesis of XOS of different lengths (four, six, eight, and ten xylose units) was confirmed by ESI-MS (Figure 4). This XOS reaction was also monitored by Nuclear Magnetic Resonance (NMR) directly while it progressed in the NMR tube. The spectra of the reaction mixture in the different time points, when compared with the spectrum of a commercial 1,4-β-d-xylotetraose, corroborate the β-1,4 regioselectivity of rXynSOS-E236G in XOS synthesis (Figure S9). Unlike the XOS generated from xylan hydrolysis, which usually contain ramifications, the enzymatic oligomerization of X2F gives rise to linear chain XOS. This type of XOS can be of biotechnological interest, because they are not frequent in nature [19].
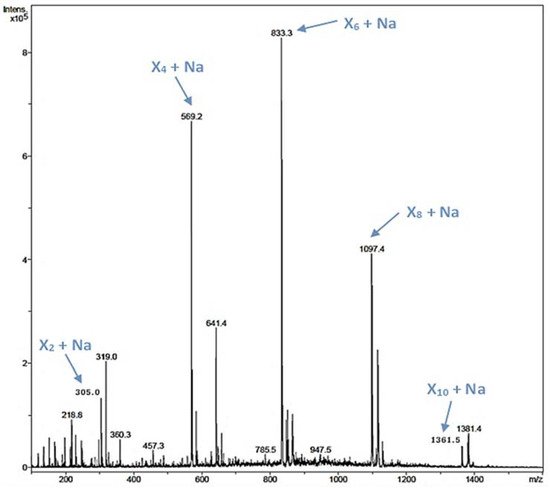
Figure 4. ESI-MS analysis of a reaction containing 20 mM X2F as the only reactant molecule and the glycosynthase variant rXynSOS-E236G as the catalyst. The m/z of ions corresponding to the Na adducts of xylooligosaccharides (XOS) of four, six, eight, and 10 xylose units (X4, X6, X8, and X10) are indicated by blue arrows.
Previous reports illustrate XOS production by the enzymatic hydrolysis of xylan using, for instance, the GH11 endoxylanase XynM from T. amestolkiae [12] and the endoxylanases BLf1 from Aspergillus brasiliensis, rXynC from P. funiculosum [46], rPoXyn3 from Penicillium occitanis [47], and rT-XynC(122)C(166) from Talaromyces thermophilus [48]. However, the application of glycosynthases to produce XOS is a promising solution, since these enzymes do not hydrolyze the XOS obtained, which increases the efficiency of the reactions [18]. Although XOS synthesis from X2F has also been described for bacterial glycosynthases, such as those from the GH10 endoxylanases XylB from Thermotoga maritima, XynB from Clostridium stercorarium, XynA from Bacillus halodurans, and Cex from Cellulomonas fimi [42,43], rXynSOS-E236G is the first fungal glycosynthase reported with this capacity.
In order to check if the glycosynthase variant rXynSOS-E236G could synthesize a wider spectrum of oligosaccharides and also to verify that other sugar donors could be employed, several reactions using XF, GF, and G2F as xylose, glucose, and cellobiose donors, respectively, were carried out. ESI-MS results confirmed the formation of XOS using XF as a donor (Table S1), but of shorter lengths (two, three, and four xylose units) and less efficiently compared to the ones obtained with X2F. The reactions with the glucose-based donors were also less efficient, as no oligosaccharides were detected with GF, and only cellotetraose was observed when employing G2F. These findings are consistent with the hydrolytic efficiencies displayed in Table 1 for rXynSOS against pNPX, pNPG and pNPG2, in which pNPX2 was clearly preferred over them.
7. Glycosylation Profile of the Glycosynthase Variant rXynSOS-E236G
The same molecules tested with the native rXynSOS were used as acceptors for the glycosynthase variant E236G (vanillyl alcohol, 2-hydroxybenzyl alcohol, hydroquinone, and gallic acid). This time, potential glycosides were clearly identified by TLC for all the phenolic compounds assayed (Figure 5a–d), and their molecular masses as sodium adducts were confirmed by ESI-MS. Table 3 shows the molecular mass of the xylobiosides detected in these reactions, although small peaks corresponding to xylotetraosides were also observed. In this regard, as all the acceptors tested have more than one hydroxyl group to potentially participate in the glycosylation reaction, it could be possible to have different xylobiosides as well as glycosides with one xylotetraose chain or two xylobiose chains located in different positions giving the same molecular mass. Overall, the results demonstrate that the glycosynthase variant rXynSOS-E236G is much more efficient than the native enzyme in glycosylation reactions.
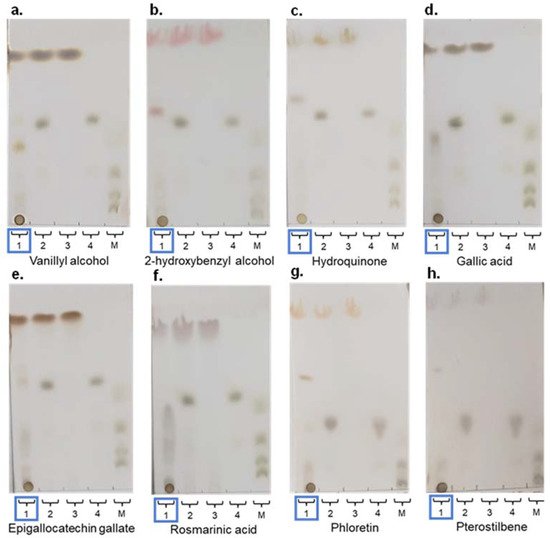
Figure 5. Thin layer chromatography (TLC) analysis of glycosylation reactions catalyzed by rXynSOS-E236G, using 20 mM X2F as donor and different acceptor molecules at 20 mM: (a) vanillyl alcohol; (b) 2-hydroxybenzyl alcohol; (c) hydroquinone; (d) gallic acid; (e) epigallocatechin gallate (EGCG); (f) rosmarinic acid; (g) phloretin; and (h) pterostilbene. Lane 1 (highlighted with a blue rectangle): samples of glycosylation mixtures containing the acceptor indicated in every case, X2F as xylobiose donor, the catalyst, and the reaction products. Lane 2: negative control with acceptor and X2F and no catalyst. Lane 3: negative control containing only the acceptor molecule. Lane 4: negative control containing only the donor X2F. Lane 5. Standards mixture with xylose, xylobiose, xylotriose, and xylotetraose.
Table 3. Synthesis of glycosides by the glycosynthase variant rXynSOS-E236G using 20 mM X2F as xylobiose donor and different acceptor molecules at 20 mM. Glycosides were detected by ESI-MS in the positive mode as Na-adducts. The yield of the main glycoside of each reaction was calculated by HPLC.
| Glycoside | Mass ESI-MS (m/z) 1 | Yield HPLC (%) |
|---|---|---|
| Vanillyl alcohol-X2 | 441.1 | 4.3 |
| 2-hydroxybenzyl alcohol-X2 | 411.0 | 7.6 |
| Hydroquinone-X2 | 397.0 | 26.0 |
| Gallic acid-X2 | 457.1 | 21.7 |
| Epigallocatechin gallate (EGCG)-X2 | 745.2 | 1.8 |
| Rosmarinic acid-X2 (two main glycosides detected) |
647.2 | 20.9 (1) 20.0 (2) |
| Phloretin-X2 | 561.1 | 7.8 |
| Pterostilbene-X2 | 543.3 | 1.6 |
1 Detected as Na-adducts.
In addition, more complex molecules such as the polyphenols epigallocatechin gallate (EGCG), rosmarinic acid, phloretin, and pterostilbene (Figure S4) were assayed as possible rXynSOS-E236G acceptors using X2F as the donor. These polyphenols are extracted from plants and exhibit numerous bioactive properties, including antioxidant, antihypertensive, antitumoral, bactericidal, neuroprotective, and anti-inflammatory activities [49]. Potential glycosides were detected by TLC for all these polyphenols (Figure 5e–h), and ESI-MS studies of their reaction products indicated the presence of different glycosides harboring xylobiose (Table 3) and others containing xylotetraose.
To further study these glycoside mixtures, HPLC analyses were carried out. One major peak as the main glycosylated product was observed for all the acceptors except for rosmarinic acid, for which two main xylobiosides were noticed (data not shown). Additionally, minor peaks were also detected, probably corresponding to glycosides containing the xylobiose in a different position or even a xylotetraose moiety. For phloretin, pterostilbene, and especially rosmarinic acid, more minor peaks and thereby wider glycoside mixtures were obtained as the result of using more complex acceptor molecules that include several potentially glycosylable hydroxyl groups.
The yield of the main glycoside in each reaction relative to the initial acceptor concentration was also calculated from the HPLC chromatograms, achieving the highest values with hydroquinone (26.0%), gallic acid (21.7%), and rosmarinic acid (two major peaks of 20.9% and 20.0%, which represents a 40.9% acceptor conversion) (Table 3). However, it must be noted that in the case of phloretin and pterostilbene, some precipitation of the acceptor molecules was observed during the reactions due to their low solubility under the conditions tested [50,51], which could have caused an underestimation of glycoside yields. These results confirm that rXynSOS-E236G glycosynthase is a promising tool to glycosylate a broad range of phenolic compounds with biotechnological applications. Furthermore, the high-glycoside yields obtained with some of the acceptors assayed could be further increased through the optimization of reaction conditions by, for instance, using multiparametric models that could determine the best values for the reaction parameters (i.e., acceptor, donor, and enzyme concentrations, time, etc.) [29,52,53].
The structure and regioselectivity of the main xylobiosides obtained in the reactions with vanillyl alcohol, phloretin, and rosmarinic acid were determined by NMR. These glycosides were selected based on their high yields, ease to purify, and biotechnological interest. Their deduced glycoside structures are depicted in Figure 6 and the corresponding chemical shifts.
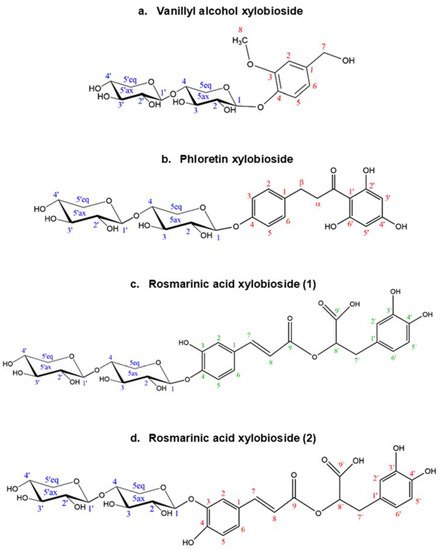
Figure 6. Structures deduced from NMR analysis of the glycosides of (a) vanillyl alcohol, (b) phloretin, and (c,d) rosmarinic acid. Every C atom in the molecules is numbered to clarify the identification of the signals.
The glycosylation of phenolic antioxidants is of biotechnological interest, as it has been shown to increase their solubility, which may lead to a higher bioavailability and better bioactive properties [54]. In this sense, previous studies have described the glucosylation of some of the phenolic compounds tested in this work and its positive effect on their properties. For instance, the glucoside of pterostilbene was proven to improve the solubility of the original aglycone and to reduce its toxicity for several human cell lines [49], while the glucosylation of phloretin also led to a higher solubility and a lower skin penetrability, which could favor a prolonged protection of the external skin layers by cosmetic preparations [55]. Nevertheless, very few cellobiosides of phenolic compounds obtained through enzymatic synthesis can be found. Still, this type of glycosylation was demonstrated to significantly increase the solubility and stability of hydroquinone, methyl gallate, ethyl gallate, propyl gallate, and epicatechin [56]. The enzymatic synthesis and applications of xylosides of phenolic compounds have also received less attention compared with the glucoside ones. Among the published data, the xyloside of hydroxytyrosol produced by the β-xylosidase BxTW1 from T. amestolkiae showed an enhancement of its neuroprotective capacity and antioxidant activity [57]. The thioglycoligase derived from the enzyme used in this last study was also able to xylosylate a broad range of phenolic compounds including gallic acid, EGCG, phloretin, and pterostilbene [58]. Regarding xylobiosides of phenolic compounds, very little is known about their enzymatic synthesis and improved biological properties other than being a dihydroresveratrol xylobioside produced by chemical synthesis and acting as an effective melanogenesis activator [59].
This entry is adapted from the peer-reviewed paper 10.3390/ijms23031383
This entry is offline, you can click here to edit this entry!
