Cd is a naturally occurring environmental toxicant, which is easily absorbed and accumulated by plants, and has strong teratogenic and mutagenic effects on organisms. Human exposure to Cd for a long time can easily cause diseases such as osteoporosis and kidney damage. It is also positively correlated with the outbreak of a variety of cancers, and is classified as a human carcinogen by the International Agency for Research on Cancer (Group I). A large amount of data shows that the intake of Cd in the human body is mainly from the diet. Therefore, it is very important to understand the influx and transport mechanism of Cd in plants.
1. Introduction
Cd is a naturally occurring environmental toxicant, which is easily absorbed and accumulated by plants, and has strong teratogenic and mutagenic effects on organisms. Human exposure to Cd for a long time can easily cause diseases such as osteoporosis
[1] and kidney damage
[2]. It is also positively correlated with the outbreak of a variety of cancers
[3], and is classified as a human carcinogen by the International Agency for Research on Cancer (Group I)
[4]. A large amount of data shows that the intake of Cd in the human body is mainly from the diet
[5]. Therefore, it is very important to understand the influx and transport mechanism of Cd in plants.
The Cd enters the soil through man-made activities such as Zn mining, smelting, application of fertilizers and pesticides
[6]. When the content of Cd in the soil is too high, it will inhibit the influx of iron by plants, causing plant growth retardation, chlorosis and other symptoms similar to iron deficiency
[7][8]. These symptoms seem to be caused by the direct or indirect interaction between Cd and Fe. Cd toxicity may also cause phosphorus deficiency or reduce manganese input, and interfere with the influx and transport of several essential nutrients (such as calcium, magnesium, phosphorus and potassium) and water by plants
[9]. In summary, Cd can enter the cell through essential element transporters. Due to the lack of specificity of these transporters, Cd, as a non-essential element, is usually assumed to be absorbed by transporters of essential elements (such as Zn, Fe, and Ca)
[10][11]. Most of the early literature also pointed out that the transporter-mediated symbiotic and the coupled transcellular pathway is the only way for Cd to flow in
[12][13]. The members of the natural resistance-associated macrophage protein (NRAMP), ZRT, IRT-like protein (ZIP), metallothionein (MT) and plant defensins (PDF), cation/proton exchanger (CAX), ATP-binding cassette transporters (ABC), metal tolerance protein/cation diffusion facilitator (MTP/CDF), heavy metal ATPase (HMA), oligopeptide transporter (OPT) and plant Cd resistance (PCR) families are closely related to the influx, chelation, transport and efflux of Cd in plants.
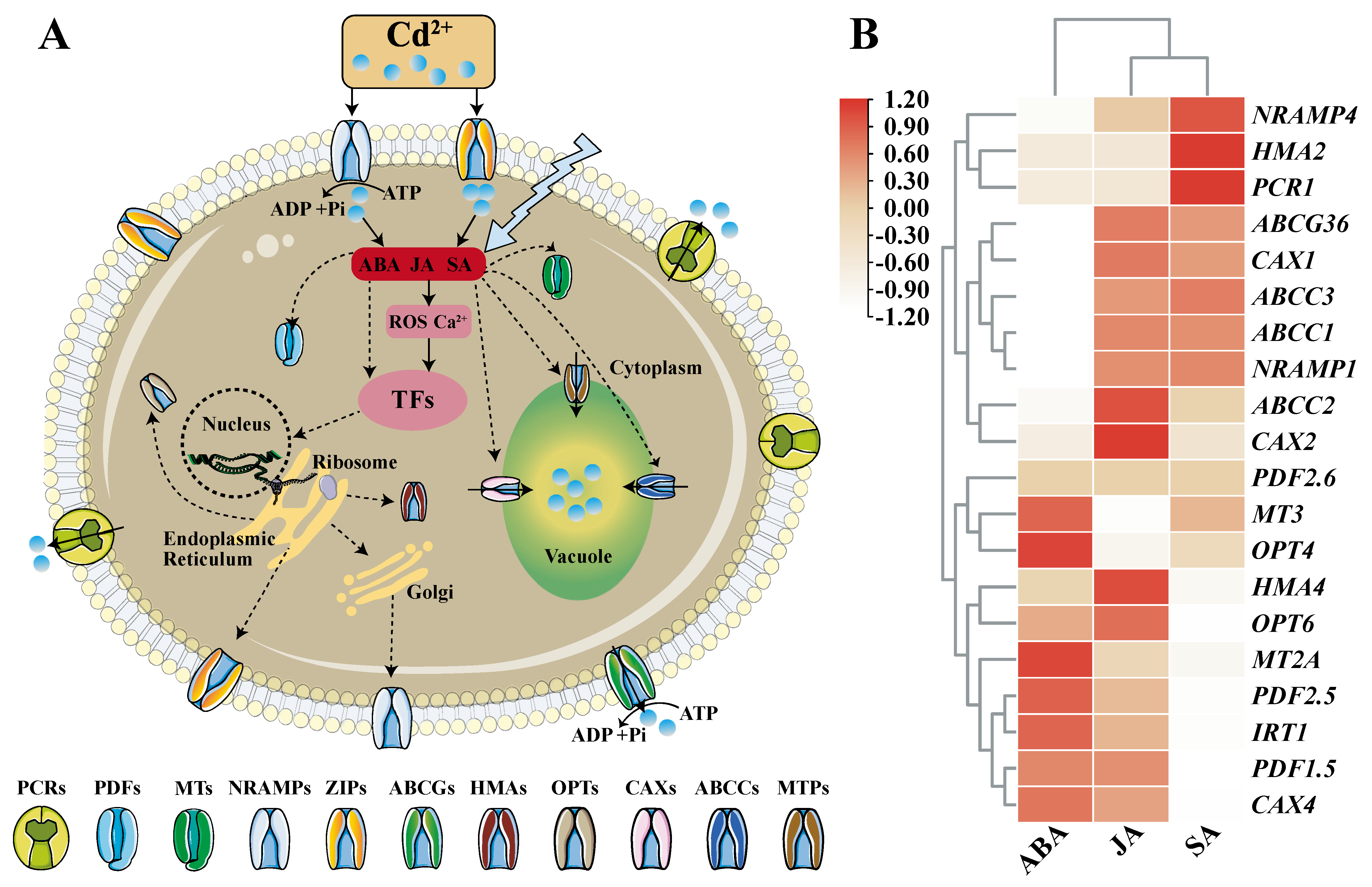
When plants are stressed by Cd, plant hormones such as abscisic acid (ABA), jasmonic acid (JA), and salicylic acid (SA) will increase and have an anti-stress effect. It was reported that exogenous application of growth regulators like ABA, JA and SA, chemical regulators and nutrient management (application of phosphorus, silicon, etc.) can improve plant tolerance to cadmium
[14]. By integrating the data in
http://ipf.sustech.edu.cn/pub/athrna/ (accessed on 30 December 2021), it was found that ABA, JA, SA hormones are closely related to the expression of cadmium transport and detoxification genes (
Figure 1). Studies on their mechanism of action have found that they can act as signaling factors to up-regulate the expression of cadmium-tolerant protein genes or down-regulate the synthesis of absorption and transport proteins, thereby improving the detoxification ability of plants to cadmium (
Figure 1)
[15]. For example, ABA can inhibit the expression of Cd uptake genes and lower Cd accumulation
[16], and SA significantly reduces Cd in rice grains by regulating the expression levels of genes associated with Cd translocation and accumulation, such as
OsNRAMP2 and
OsHMA3 [17]. JA inhibits the expression of genes that promote Cd uptake and long-distance translocation, positively limits Cd accumulation and alleviates Cd toxicity in Arabidopsis through the jasmine acid signaling pathway
[18]. Similarly, the significant effect of Si in reducing Cd accumulation has also been attributed to its down-regulation of the expression of genes involved in Cd uptake (
OsNramp5) and root-to-shoot translocation (
OsHMA2)
[19]. Exogenous addition of astaxanthin and its gold nanoparticles
[20], β-cyclocitral
[21], glutamic acid
[22], nitric oxide
[23], etc. increased the tolerance of plants to Cd stress, which is also because they alter the expression of MTP, HMA, OPT, ZIP, NRAMP, ABC and other family members. In the main, the influx, transport and detoxification mechanism of Cd by plants are closely related to proteins.
Figure 1. Plant hormone signaling regulation and related gene expression analysis under Cd stress. A: The regulatory pathway of plant hormones under Cd stress. Plant hormones such as ABA, JA, SA enhance the detoxification ability of plants against cadmium stress by regulating the expression of downstream proteins. TFs stands for transcription factors. The arrows with solid lines indicate that the regulation methods have been reported so far, and the arrows with dotted lines indicate that the regulation methods are not yet clear. Polyline arrows indicate exogenous additions. B: Effects of ABA, JA and SA on the expression of genes involved in Cd transport and detoxification (taking Arabidopsis as an example). Expression data comes from the database (
http://ipf.sustech.edu.cn/pub/athrna/) (accessed on 30 December 2021). A red box means more correlation, a white box means less correlation. Plant hormones like ABA, JA, SA are closely related to the expression of these genes.
As so often is the case, the realization of biological functions depends on the interaction between ligand binding residues and metal ions, and the molecular mechanism involves the binding of metal ions to specific residues in the protein. Some of these amino acid motifs are conserved in the protein family, and they can produce key stable interactions or play important functional roles
[24]. Each family has its own conserved motifs, which play an important role in protein recognition and metal ion binding.
2. Influx Protein
There are two different processes in the influx of Cd by plant roots. The first is a temperature-independent process, which may be passive, reaching an equilibrium concentration after a few seconds or minutes. The second is the physiological process, which mainly depends on the relevant transport proteins in plants to play a role
[25]. There is no Cd selective transporter in plant cells. Cd in soil enters plant cells mainly through plasma membrane transporters such as NRAMP and ZIP
[26].
2.1. A Brief Introduction to the NRAMP Family, the Key Proteins Related to Cd Influx and the Mechanism of Action
NRAMP, a proton-coupled metal ion transporter, is an ancient intact membrane transporter family. It is widely present in bacteria, fungi, plants and animals, and participates in the transportation of a variety of divalent metal ions
[27]. Using online software (
http://wlab.ethz.ch/protter/start/) (accessed on 30 December 2021) to map their transmembrane domains (TMDs) and amino acid structural features
[28], it was found that they typically have 12 TMDs. The DPGN residue near TMD1 was identified as the binding site of the NRAMP transporter, which coordinates with divalent metal ions and participates in the transport of Cd to plant root cells
[29]. Mutations in these residues can cause impaired transporter function. In Arabidopsis, a total of 6 NRAMP members have been identified, of which AtNRAMP1, AtNRAMP3 and AtNRAMP4 are related to the influx and transport of Cd
[30]. Yeasts and Arabidopsis that overexpress these genes can increase their sensitivity to Cd and contain more Cd in yeast cells. There were seven NRAMP genes found in rice. Among these members, OsNramp1 and OsNramp5 were identified as the main Cd uptake protein in rice
[31]. It is interesting that even in the same species, NRAMP has undergone some interesting changes to regulate the accumulation of heavy metals. For example, the amino acid sequence of OsNRAMP1 in high and low accumulation rice varieties is exactly the same, but the Cd content can be reduced by regulating the low expression of OsNRAMP1 in the roots
[32]. The transport function of NRAMP family members is driven by H co-transport, H provides energy for active transport, and the conformational changes of proteins are crucial
[33]. Residue phenylalanine413 in AtNRAMP4 is located in the highly conserved protein domain of plant and animal NRAMP members, and its mutation can change the accumulation of Cd in plants
[34]. The mutations of histidine in AtNRAMP3 severely impaired the transport of Fe and Mn, and partially impaired the transport of Cd
[35].
2.2. A Brief Introduction to the ZIP Family, the Key Proteins Related to Cd Influx and the Mechanism of Action
The ZIP family mainly comes from dicotyledonous plants, which play a major role in the homeostasis of zinc and metal in plants
[36]. They are not highly selective and can transport different metal divalent cations, including essential elements (such as Zn, Mn, Fe) and toxic elements (Cd)
[37]. The iron-regulated transporter 1 (IRT1) is the main root transporter that absorbs iron from the soil, and is also the main entry route for potentially toxic metals such as manganese, zinc, cobalt, and Cd in plants
[38][39]. For example, AtIRT1 targets the plasma membrane in the body, which can mediate the influx of Cd from the environment
[40]. When AtIRT1, OsZIP1 and OsZIP3 are expressed in yeast cells, these three genes increase the sensitivity of yeast to Cd stress and the accumulation of Cd
[41]. In rice, overexpression of OsIRT1 and OsIRT2 made cells more sensitive to Cd and increased Cd accumulation
[42]. OsZIP6 can transport at least three transition metal ions such as Fe, Co and Cd
[43]. Studies have found that most ZIP proteins have 8 potential TMDs and a similar membrane topology, and the amino and carboxyl terminal ends of the protein are located on the outer surface of the plasma membrane
[44]. The length of the ZIP protein varies from 309 to 476 amino acids
[45]. The most conserved part of the ZIP family proteins appears in TMD IV, which is expected to form an amphipathic helix with completely conserved histidine residues. The histidine residue, together with the adjacent (semi) polar residues, can form part of the heavy metal binding site in the membrane as a transport pathway
[46]. Understanding the mechanism of ZIP transporters requires their high-resolution crystal structure, which is not yet available for plant ZIP protein. However, it was found in
Bordetella bronchiseptica that the crystal structure of the BbZIP transporter has 8 TMDs and a binuclear metal center (M1, M2) was observed
[47]. M1 may be located just on the transportation route, which can quickly combine and release metals during transportation, which is necessary for the transportation of metal ions, and M2 may play a supporting role
[48]. Compared with BbZIP, plant ZIP transporters may have partially overlapping but different metal transport mechanisms, because some residues of ZIP involved in metal binding and transport are not conserved in plants
[49].
2.3. Comparison of Core Characteristics of Cd Influx Key Proteins
Compared to the key proteins for Cd uptake in rice and Arabidopsis, it was found that these proteins were mainly expressed in plant roots and located on the cell membrane, indicating that the free Cd in the soil solution was mainly transported into the plant through root transporters (
Table 1). The NRAMP protein mainly relies on H co-transport drive to provide energy, while the transport mechanism of ZIP is currently unclear. In particular, there are only about 5 NRAMP members whose evolution tends to be conservative in plants, and there is no large-scale expansion. DPGN residues are only found in NRAMP, which may be a key and special function residue. In animals, members of the NRAMP family have been shown to play an important role in the immune response, usually in the phagocytosis and presentation of antigens by pathogens, but whether there are similar functions in plants has not been reported. In addition to the Cd stress response, it seems likely that it participates in a series of foreign invasion defenses. However, there are many members in the ZIP family and their evolution has undergone specific changes. For example, all of the ZIP proteins involved in Cd influx have a signal peptide at the N-terminus. Signal peptides can play a role in controlling protein secretion rate, determining protein folding state, affecting downstream transmembrane behavior and N-terminal glycosylation, and nuclear localization signals
[50]. The changes may lead to differences in subcellular functions between them, and may produce specific stress responses to different metal ions. Between TMD3 and TMD4 of the ZIP protein, there is a histidine-rich residue (HX)n (X = any amino acid, n = 3–6) that can act as a Zn buffer bag and a cytoplasmic surface zinc level sensor, and it may be a potential metal (Zn, Co and/or Cd) binding domain. This residue is also ubiquitous in the MTP family, but surprisingly, the MTP principally produces an effect in the vacuole
[51]. Interestingly, OsZIP1 can also be located in the endoplasmic reticulum, while OsZIP3 can be expressed in xylem parenchyma cells. The parenchyma of the xylem of plant roots is the main “controller” of Cd loading into the xylem and its transport to the branches
[52], indicating that the functions of the family members have diversified. However, these changes may benefit from the emergence of the N-terminal specific signal peptide of the ZIP family. Future research should aim to determine whether it will play a more unique role, for example, the special signal peptide may be improved to become a part of the NRAMP family.
Table 1. Summary of tissue structure-specific expression, structural characteristics and functions of key Cd influx proteins.
| The Key Proteins |
Tissue Specific Expression |
Subcellular Localization |
Structural Features |
References |
| AtNRAMP1 |
Mainly expressed in the root |
Cell membrane |
12 TMDs, DPGN near to TMD1. |
[53][54] |
| OsNRAMP1 |
Mainly expressed in the mature zone of the root |
Cell membrane |
12 TMDs, DPGN near to TMD1. |
[32][55] |
| OsNRAMP5 |
Mainly expressed in the root |
Cell membrane |
10 TMDs, DPGN is near to TMD1. There is a Asparticacid residue (DXXXD, X= any amino acid) at the C-terminus. |
[31] |
| AtIRT1 |
Root epidermal cells |
Cell membrane |
8 TMDs; there is a signal peptide at the N-terminus, histidine, Asparticacid residues (HXXXD, X= any amino acid) on TMD2, and (HX)n between TMD3 and TMD4. |
[40] |
| OsIRT1 |
Mainly expressed in the root |
Cell membrane |
8 TMDs; there is a signal peptide at the N-terminus, HXXXD residues on TMD2, and the (HX)n residues between TMD3 and TMD4. |
[56] |
| OsIRT2 |
Mainly expressed in the root |
Cell membrane |
There are 8 TMDs with signal peptide and DXXXD residues at the N-terminus; the (HX)n residues between TMD3-TMD4. |
[56] |
| OsZIP1 |
Mainly expressed in the root |
Cell membrane |
8 TMDs, with signal peptide and DXXXD residues at the N-terminus; HXXXD residues on TMD2, and the (HX)n residues between TMD3-TMD4. |
[41] |
| OsZIP3 |
Xylem parenchyma cells |
Cell membrane |
There are 7 TMDs, there is a signal peptide at the N-terminus, and there are (HX)n residues and HXXXD conserved residues between TMD3-TMD4. |
[41][57] |
3. Chelating Protein
Only a small part of the metal content in plants exists in the form of free water ions, and most of the ions are combined with low molecular weight ligands or proteins. The cell wall is the first protective barrier to prevent heavy metals from invading cells, the first “living” structure, and the target of heavy metal damage. The immobilization of Cd in the cell wall participates in the tolerance mechanism of plants to Cd, and PDF exists in the cell wall and can participate in the chelation of it. However, MT mainly exists in the cytoplasm and can combine with free Cd to reduce the harm of heavy metals to plants.
3.1. A Brief Introduction to the MT Family, Key Proteins and Chelation Mechanism
MT is a low-molecular-weight metalloprotein in cells that is rich in cysteine and exists in all organisms except archaea
[58]. According to the distribution pattern of cysteine, it can be divided into 4 sub-families: MT1, MT2, MT3 and MT4
[59]. The MT1-3 subfamily has two cysteine-rich regions, 6 cysteines in the C-terminal domain, and 6, 8 and 4 cysteines in the N-terminal cysteine-rich region, respectively. M4 has three cysteine-rich regions, each containing 6, 6 and 5 cysteine residues
[60]. It can be combined with heavy metal Cd with high affinity, and plays a role in detoxification of heavy metals and maintaining the stable state of essential metal ions in cells. Among them, MT1, because of its unique metal bonding characteristics, can endow Cd resistance and contribute to the stable state of zinc
[61]. The heterologous expression of OsMTI-1b can improve the tolerance of
E. coli cells to Cd by chelating metal ions
[62]. Overexpression of OsMT1e improved rice growth under Cd stress
[63]. After CdCl
2 treatment, Arabidopsis lines overexpressing OsMT-3a accumulated higher levels of Cd in shoots and roots
[64]. We can see that the primary structure of MT is obviously rich in highly conserved cysteine residues such as CC, CXC and CXXC (X = any amino acid). These residues have the remarkable ability to bind a large number of monovalent or divalent metal ions, as well as maintain the steady-state of basic metals and the ability to detoxify (toxic) metals, especially copper, zinc and Cd
[65]. The sulfhydryl groups on MTs can combine with heavy metal ions to form non-toxic or low-toxic compounds, thereby eliminating the toxicity of heavy metals
[66]. Furthermore, the mercaptans (esters) in MTs can act as powerful antioxidants. They usually bind metal ions in metal thiol clusters to provide high thermodynamic stability and kinetic instability
[67]. MT constitutes a highly complex system, even though the same peptide can show different final 3D folds, it is expected that these folds are related to different functions
[68].
3.2. A Brief Introduction to the PDF Family, Key Proteins and Chelation Mechanism
PDF is a type of small molecule protein, each containing 45 to 54 amino acids, and 13 members have been identified in Arabidopsis
[69]. PDF can be divided into two families, namely PDF1 and PDF2, which mainly play a role in fungal resistance and zinc tolerance, but it is interesting that the recent reports in the literature show that they can participate in the chelation of Cd. For instance, the PDF2 (AtPDF2.5) in Arabidopsis is involved in Cd tolerance and accumulation, and Cd binding analysis shows that AtPDF2.5 has Cd chelating activity in vitro
[70]. Overexpression of AtPDF2.6 increases the tolerance of
E. coli and yeast to Cd, and the in vitro Cd binding test shows that AtPDF2.6 also has Cd chelating activity
[71]. The PDF1.5 is located in the cell wall and may detoxify Cd by chelating Cd to the cell wall
[61]. PDF has a characteristic three-dimensional folding pattern, which is stabilized by eight disulfide bonds between cysteines
[72]. Cysteine and other key residues are necessary to induce changes in the three-dimensional structure of AtPDF, which can mediate and increase yeast tolerance to Cd
[73].
3.3. Comparison of the Core Features of Key Cd Chelation Proteins
Analyzing the proteins involved in Cd chelation in the MT and PDF families showed that these proteins all have a cysteine-rich domain (
Table 2). Cysteine is a sulfur-containing amino acid with physiological activity, with amino and sulfhydryl groups. Among protein amino acids, cysteine may be the only amino acid that contains a thiol group on the side chain and has a binding site for heavy metals of plant cells
[74]. PDF can chelate Cd in the cell wall, thereby preventing Cd from destroying the cell structure, and has a more effective detoxification ability. Nevertheless, MT forms a chelate with Cd in the cytoplasm, and is then transferred to the vacuole by other transporters for detoxification. Interestingly, there is a special signal peptide at the N-terminus of the PDF protein, which may mediate its secretion in the cell wall. Surprisingly, OsMTI-1b and OsMT1e are expressed in the nucleus, indicating that members of the MT family seem to be involved in the regulation of gene transcription. On the whole, the cell wall is a unique cell structure of plants, and it is the first protective barrier to prevent heavy metals from invading cells. The specific signal peptide at the N-terminus of PDF protein has great research value for the genetic improvement of crop Cd tolerance, and how the MT family members regulate gene expression in the nucleus remains to be explored.
Table 2. The tissue structure specific expression, structure characteristics and function summary of the key proteins of Cd chelation.
| The Key Proteins |
Tissue Specific Expression |
Subcellular Localization |
Structural Features |
References |
| AtMT2a |
/ |
Cytoplasm |
CXCXXXCXC and DXXXD are distributed at the C end, without TMDs. |
[75] |
| AtMT3 |
/ |
Cytoplasm |
CXCXXXCXCXXXCXC is distributed at the C-terminus and has no TMDs. |
[75][76] |
| OsMTI-1b |
/ |
Cytoplasm, Nucleus |
CXCXXXCXCXXXCXC is distributed at the C-terminus and has no TMDs. |
[62] |
| OsMTI-2b |
Highly expressed in rice stems |
Cytoplasm |
N-terminal has 8 cysteine residues arranged in CC, CXC and CXXC residues. |
[77] |
| OsMT1e |
Expressed in roots at all developmental stages |
Nucleus |
CXCXXXCXCXXCXCXCXCX is distributed at the N-terminal and has no TMDs. |
[78] |
| OsMT-3a |
/ |
Cytoplasm |
CXCXXXCXCXXCXCXCXCX is distributed at the C-terminus and has no TMDs. |
[59][64] |
| AtPDF1.5 |
Mainly expressed in knots and peels |
Cell wall and cytoplasm |
There is a signal peptide at the N-terminus, a cysteine-rich domain, and a DXXXD residue. |
[72] |
| AtPDF2.5 |
mainly expressed in root vascular bundle |
Cell wall |
Cysteine-rich domain and secretion signal peptide. |
[70] |
| AtPDF2.6 |
mainly expressed in root vascular bundle |
Cell wall |
There is a signal peptide at the N-terminus, a cysteine-rich domain. |
[71] |
4. Vacuolar Protein
The cellular mechanism of Cd is generally considered to include isolation in the cytoplasm by glutathione, glutathione-related plant chelator, and isolation in the vacuole by the vacuolar membrane Cd/H antiporter. In fact, most plants detoxify heavy metals by storing Cd-chelating complexes or free Cd in vacuoles, because this organelle contains enzymes involved in Cd detoxification, such as phosphatase, lipase, and protease. The main proteins involved in the transfer of Cd into vacuoles in plants are CAX, ABCC and MTP family members.
4.1. Introduction to CAX Family, Functional Proteins and Vacuolar Transfer Mechanism
CAX is a secondary charged ion transporter located in the vacuole, which relies on the secondary energy transport of metal ions in the proton transmembrane gradient
[79]. The main function of CAX protein is to limit the toxicity of certain free metal ions to cells, such as Mn or Cd, provide tolerance to metal toxicity, and maintain cytosolic concentration
[80]. For example, the expression of AtCAX2 in tobacco increases the transport of Cd in root vacuoles
[81]. AtCAX4 compensates to a certain extent for the reduction of the vacuolar membrane H pump and the increase in proton leakage, which helps to improve the higher tolerance of plants to Cd
[82]. OsCAX1a/OsCAX1c may also be involved in the process of Cd transfer to vacuole storage
[83]. In plants, CAX is driven by the pH gradient produced by vacuolar H-ATPase and H-pyrophosphatase to drive metal ion transport. Studies have shown that the local primary structure determines the cation specificity of CAX binding
[84]. There is a highly conserved 36 motif region between TM3 and TM4, TM8 and TM9, named c-1 loop and c-2 loop, respectively, which may form a vestibule or filter for cation selection, and there are highly conserved GNXXE residues
[85]. CAX has a common motif, which is the region rich in acidic amino acid motifs between TM6 and TM7, which is believed to be related to the capture and selection of metal ions.
4.2. An Introduction to the ABCC Subfamily, Functional Proteins and Vacuolar Transfer Mechanism
The ABC transporter is one of the largest and oldest protein families; it plays a very crucial part in plant heavy metal transport and plant disease resistance. A total of 9 subfamilies (ABCA-ABCI) have been identified in eukaryotes, of which 8 subfamilies (ABCA-ABCG and ABCI) exist in the plant genome. In plants, the two subfamilies that are currently reported to be related to heavy metals are mainly ABCC and ABCG. ABCC mainly transfers Cd to vacuoles, while ABCG removes Cd from cells for detoxification. The AtABCC1 and AtABCC2 participate in the vacuolar separation of PC-Cd (II) and PC-Hg (II), thereby making plants tolerant to these toxic heavy metals
[86]. AtABCC3 detoxifies by transporting the PC-Cd complex into the vacuole
[87], and the OsABCC9 isolates Cd into the vacuoles of rice roots to mediate Cd tolerance and accumulation
[88]. They are usually composed of two membrane domains or TMDs and two cytoplasmic nucleotide binding domains (NBD)
[89]. Two TMDs provide channels for metal ions, while two NBDs combine and hydrolyze ATP to provide energy for transport reactions
[90]. As the input proteins, TMD and NBD act as separate polypeptide chains, while in the bacterial export protein, a TMD and NBD are fused to form a homodimer or a heterodimer, thereby producing a complete and functional transport protein
[91]. In particular, high-resolution electron microscopy has determined the structure of 8 different states and conformations, including two inward (IF), four outward (OF) and two asymmetric hydrolyzed states
[92]. A single conformational transition from IF to OF conformation, triggered by ATP binding, drives unidirectional substrate translocation across the membrane. After this power stroke, ATP hydrolysis and phosphate release start to return to a static state, which promotes nucleotide exchange and a new round of substrate binding and translocation
[93].
4.3. Introduction to MTP Family, Functional Proteins and Vacuolar Transfer Mechanism
CDF is also known as MTP in plants. MTP/CDF is a type of membrane-bound protein, which can maintain cell homeostasis and play a vital part in the process of plants dealing with heavy metal stress. It is a ubiquitous divalent cation transporter and is essential for the metal homeostasis and tolerance of archaea, bacteria and eukaryotes. Most of the CDF family members confer heavy metal tolerance by affecting the outflow of heavy metals from the cytoplasm. Due to the different members, outflow refers to the outflow from the cell, or into the internal compartment
[94][95]. In the model plants rice and Arabidopsis, members of the MTP family mainly transfer heavy metals into vacuoles to increase plant tolerance. For instance, heterologous expression of OsMTP1 in tobacco plants increases the plant’s tolerance to Cd toxicity and allows transgenic plants to accumulate more Cd
[96]. After 24 h of Cd stress, the expression levels of OsMTP6, OsMTP7, OsMTP9 and MTP11.1 in the shoots of rice increased significantly
[90], but there is no research showing how they respond to Cd stress
[97]. All CDF transporters consist of two domains: the TMD for cation transport and the regulatory cytoplasmic C-terminal domain (CTD)
[98]. The combination of metal ions and CTD will induce its dimerization, and it will lead to a higher CTD accumulation
[99]. Although CTD shows a high degree of sequence variability between different species, all available CTD structures have similar metal chaperone-like folds
[100]. Metal chaperones are cytoplasmic metal carrier proteins that form a metal donor–acceptor interface with related transport mechanisms to transport metal ions to various protein targets and participate in cytoplasmic metal transport
[101]. They usually have 6 TMDs and have a highly conserved characteristic sequence in TMD 2 and TMD 5, which has a potential effect on metal selectivity
[102][103]. There are also HXXXD or DXXXD conserved residues on or near these 2 TMDs. If this residue is subjected to site-directed mutation, the CDF transporter will lose its ability to bind to Cd and impair Cd transport
[104]. However, so far, the metal selectivity of the substrate among members of the CDF family is still unclear. We speculate that these two conserved and aspartic acid-rich residues are key targets for the binding and transport of metal ions. Furthermore, some MTP transporters have an (HX)n between TMD 4 and TMD 5 that also exists in members of the ZIP family and has similar functions. It can be used as a Zn buffer bag and a cytoplasmic surface zinc level that recognizes Sensor
[105].
4.4. Comparison of Core Characteristics of Key Proteins in Vacuolar Transport
Vacuoles are the largest organelles in mature plant cells. They are a reservoir of ions and metabolites and are indispensable for the detoxification process and normal cell development. By comparing the results of subcellular localization, it is found that CAX/MTP/ABCC family members are mainly located on the vacuole membrane, which will increase the content of Cd in overexpressed plants, indicating that these proteins are detoxified by transferring Cd to the vacuole storage (Table 3). H/DXXXD residues are widely distributed in these three family members, whether they are a specific Cd binding site remains to be considered. The difference is that CAX and ABCC family members can be expressed both in the vacuole membrane and in the vacuole. Whether they have multiple functions, such as chelating Cd in the vacuole and transporting on the vacuole membrane, remains to be studied in depth. The ABCC transporter in particular can not only transport Cd directly, but also transport the PC-Cd complex. Whether there is a specific theme to give this interesting substrate difference or not requires continued research.
Table 3. Tissue structure-specific expression, structural characteristics and function summary of key proteins in vacuolar transport.
| The Key Proteins |
Tissue Specific Expression |
Subcellular Localization |
Structural Features |
References |
|
| AtCAX2 |
/ |
Tonoplast |
8 TMDs; with GNXXE residue on TMD2 and TMD7. |
[81] |
|
| AtCAX4 |
Mainly expressed in the root |
Tonoplast |
10 TMDs; a GNXXE residue on TMD7, and the DXXXD residues between TMD5-6. |
[82] |
|
| OsCAX1a |
expressed at a high level in flowering spikelet |
Vacuole |
10 TMDs; GNXXE residues on TMD3 and TMD8, and the DXXXD residues between TMD6-TMD7. |
[83] |
| OsCAX1c |
Strongly expressed in leaves |
Tonoplast |
10 TMDs; GNXXE residues exist on TMD2, TMD, and DXXXD residues near TMD6. |
[83] |
| AtABCC1 |
Mainly expressed in the root |
Vacuole |
15 TMDs, nucleotide binding site at the C-terminus, cysteine-rich loop, Q-loop, Walker B residue. |
[86] |
| AtABCC2 |
Mainly expressed in the root |
Vacuole |
10 TMDs, cysteine-rich loop, Walker B residue, DXXXD residues exist between TMD4-TMD5. |
[86] |
| AtABCC3 |
/ |
Tonoplast |
15 TMDs, cysteine-rich loop, Walker B residue, HXXXD residues near TMD3, and DXXXD residues between TMD9-TMD10. |
[87] |
| OsABCC9 |
Significantly induced by Cd treatment in roots |
Tonoplast |
8 TMDs, cysteine-rich loop, Q-loop, Walker B residue, and HXXXD and DXXXD residues at the N-terminus. |
[88] |
| OsMTP1 |
Significantly induced by Cd treatment in roots |
Vacuole |
The characteristic residue of CDF (SLAILTDAAHLLSDVAA), 6 TMDs and histidine-rich regions, HXXXD residues exist on and/or near TMD2, TMD5. |
[96] |
This entry is adapted from the peer-reviewed paper 10.3390/ijms23031734