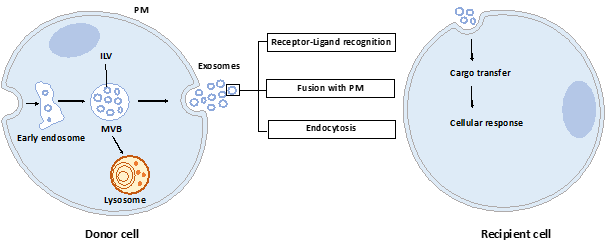
Exosomes are nano-sized vesicles generated inside cells during the maturation of endosomes into multivesicular bodies (MVBs) and then released into the extracellular space.
- exosomes
- extracellular vesicle
- lipids
- proteins
- nucleic acids
1. Introduction
Extracellular vesicles (EVs), such as exosomes, are critical mediators of intercellular communication between tumor cells and other cells located in the microenvironment but also in more distant sites. Exosomes are small EVs that can carry a variety of molecules, such as lipids, proteins, and non-coding RNA, especially microRNAs (miRNAs).
2. Exosome Biogenesis and Secretion
Exosomes are nano-sized vesicles generated inside cells during the maturation of endosomes into multivesicular bodies (MVBs). This is a multistep process starting with the invagination of the plasma membrane leading to the formation of early endosomes (Figure 1). Next, many small vesicles called intraluminal vesicles (ILVs) are formed inside them by the inward invagination of their membranes, and such endosomes are called MVBs[1]. The endosomal sorting complex required for transport (ESCRT) machinery is involved in this step. This machinery is composed of four complexes (ESCRT-0, I, II, III) working sequentially, but some evidence showed that vesicles can also be formed without them, involving for example tetraspanins[2] and/or lipids[3][4]. Finally, MVBs either fuse with lysosomes in order to hydrolyze their content, or with the plasma membrane to release the ILVs, called now exosomes, into the extracellular space. Similarly, the exosome secretion from the parent cell involves multiple steps: trafficking of MVBs to the plasma membrane, docking, fusion, and release[5].
Figure 1. Exosome biogenesis, secretion, and capture. Exosomes are formed by the inward budding of intraluminal vesicles (ILV) during the maturation of the early endosomes into the multivesicular body (MVB). ILV become exosomes after their release into the extracellular space, thus after the fusion of the MVB with the plasma membrane (PM). Released exosomes can interact with the recipient cell through receptor-ligand recognition, fusion with PM, or endocytosis. Cargo can be then transferred into the cytoplasm of the recipient cell where it can exert its effects.
Once released by parent cells, exosomes can interact with neighboring or distant cell via at least three mechanisms[6]: i) interaction between exosome transmembrane proteins and the signaling receptors of recipient cells, ii) exosome fusion with the plasma membrane of recipient cell and release of their content into the cytosol, iii) exosome internalization into the recipient cell by endocytosis. Interestingly, it has been shown that exosomal miRNAs released in the cytoplasm are functional and may induce function and phenotype changes in recipient cells[7][8].
3. Exosome Content
During their formation, specific constituents of the parent cell content are trapped into exosomes. A wide variety of molecules are thus found in exosomes: lipids, proteins, and nucleic acids. According to the present version of the exosome content databases (ExoCarta[9], Vesiclepedia[10] or EVpedia[11]), more than 9700 proteins, 1100 lipids, 3400 mRNAs and 2800 miRNAs have been identified in various exosomes or small EVs.
3.1. Lipids
The exosome membrane is enriched in cholesterol, sphingomyelin, ceramides, phosphatidylserine, phosphatidylinositol, and phosphatidic acid[12]. Their lipids’ compositions impact their membrane fluidity, allowing them to be stable in the extracellular space, but also impacting their own formation. Indeed, membrane curvature is strongly dependent on membrane lipids, themselves conditional on the enzymes available in the cell, such as the sphingomyelinase 2 (nSMase2)[4] or phospholipase D2 (PLD2)[13].
3.2. Proteins
Exosomes contain many proteins, and some of them are highly enriched, such as tetraspanins (CD63, CD81, CD9), heat shock proteins (HSP70, HSP90), MVBs formation proteins (TSG101, Alix), or others, and could therefore be used as markers for exosome characterization[14]. Interestingly, exosomes also carry immune proteins, such as those of major histocompatibility complex I and II (MHC I/II) or PD-L1 (programmed death ligand 1), and can be consequently involved in the modulation of the immune response against the tumor (see parts 4 and 5).
3.3. Nucleic acids
Exosomes are enriched in RNAs, especially in small RNA such as miRNAs. Exosomal miRNAs are not randomly incorporated into them, pointing out the existence of a mechanism for active sorting of specific miRNAs into exosomes. The exact mechanisms by which this selection is made are not totally understood. However, four potential pathways have been proposed. The nSMase2 was the first molecule to be involved in the miRNAs sorting into exosomes. In 2013, Kosaka and collaborators[15] reported that the overexpression of the nSMase2 increased the miRNAs levels in HEK293-derived exosomes and, in contrast, a decrease of its expression was associated with a lower content of miRNAs in exosomes. Another sorting mechanism was described the same year by Villarroya-Beltri et al.[16]. They described the involvement of the sumoylated version of the RNA-binding protein hnRNPA2B1 in the miRNAs sorting into T cells-derived exosomes. This protein recognizes a specific motif GAGAG called “EXO-motif” in the miRNA 3’end sequence and allows a selective loading of these miRNAs into exosomes. Since this discovery, other RNA-binding proteins including SYNCRIP (synaptotagmin binding, cytoplasmic RNA interacting protein)[17] and YBX1 (Y box binding protein 1)[18] have been identified for their role in the miRNA sorting. The third mechanism has been described by Kopper-Lalic et al. in 2014[19]. They found that 3’end uridylated miRNAs were enriched into B cell-derived exosomes, whereas 3’end adenylated endogenous miRNAs stayed in the cell. This observation shows that modifications of the miRNA 3’end portion are linked to their loading into the exosome. The fourth pathway involves the miRNA-induced silencing complex (miRISC)-related pathway: mature miRNAs can interact with assembly proteins to form the miRISC complex mainly composed of the mature miRNA, its target mRNA, GW182, and AGO2 (Argonaute2). The knockout of AGO2 decreases the sorting of some miRNAs such as miR-451, miR-150, and miR-142-3p in HEK293T cell-derived exosomes, suggesting that AGO2 could be involved in the miRNA sorting into exosomes[20]. As outlined later, these sorting mechanisms could be important to translate exosomes into therapeutic applications.
The international society for extracellular vesicles (ISEV) recommends the use of the generic term extracellular vesicle (EV) as “particles naturally released from the cell that are delimited by a lipid bilayer and cannot replicate”[14]. Exosomal vesicles are included in these EVs but need to be rigorously characterized since it is often difficult to distinguish them once they leave the cell. Moreover, during their isolation, they are frequently mixed with other vesicles including microvesicles and apoptotic bodies. The term exosomes has been extensively used in the past but tend to be progressively replaced by the more accurate term, small EVs. In our review, we will use either term according to the publication source, but notably, sometimes exosomes have not been investigated according to the MISEV guidelines, and are probably in fact a mix of EVs.
This entry is adapted from the peer-reviewed paper 10.3390/ijms21176024
References
- Marina Colombo; Graça Raposo; Clotilde Théry; Biogenesis, Secretion, and Intercellular Interactions of Exosomes and Other Extracellular Vesicles. Annual Review of Cell and Developmental Biology 2014, 30, 255-289, 10.1146/annurev-cellbio-101512-122326.
- Daniel Perez-Hernandez; Cristina Gutiérrez‐Vázquez; Inmaculada Jorge; Soraya López-Martín; Angeles Ursa; Francisco Sánchez-Madrid; Jesús Vázquez; María Yáñez-Mó; The Intracellular Interactome of Tetraspanin-enriched Microdomains Reveals Their Function as Sorting Machineries toward Exosomes*. Journal of Biological Chemistry 2013, 288, 11649-11661, 10.1074/jbc.M112.445304.
- Pollet, H.; Conrard, L.; Cloos, A.-S.; Tyteca, D. Plasma membrane lipid domains as platforms for vesicle biogenesis and shedding? Biomolecules 2018, 8, 94.
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brügger, B.; Simons, M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 2008, 319, 1244–1247.
- Roberta Palmulli; Guillaume Van Niel; To be or not to be... secreted as exosomes, a balance finely tuned by the mechanisms of biogenesis. Essays in Biochemistry 2018, 62, 177-191, 10.1042/ebc20170076.
- Kelly J McKelvey; Katie L. Powell; Anthony W. Ashton; Jonathan M. Morris; Sharon A. McCracken; Exosomes: Mechanisms of Uptake. Journal of Circulating Biomarkers 2015, 4, 7, 10.5772/61186.
- Virginie Vignard; Maureen Labbé; Nadège Marec; Gwennan André-Grégoire; Nicolas Jouand; Jean-François Fonteneau; Nathalie Labarrière; Delphine Fradin; MicroRNAs in Tumor Exosomes Drive Immune Escape in Melanoma. Cancer Immunology Research 2019, 8, 255-267, 10.1158/2326-6066.cir-19-0522.
- D. Michiel Pegtel; Katherine Cosmopoulos; David A. Thorley-Lawson; Monique A. J. Van Eijndhoven; Erik S. Hopmans; Jelle L. Lindenberg; Tanja D. De Gruijl; Thomas Würdinger; Jaap M. Middeldorp; Functional delivery of viral miRNAs via exosomes. Proceedings of the National Academy of Sciences 2010, 107, 6328-6333, 10.1073/pnas.0914843107.
- Shivakumar Keerthikumar; David Chisanga; Dinuka Ariyaratne; Haidar Al Saffar; Sushma Anand; Kening Zhao; Monisha Samuel; Mohashin Pathan; Markandeya Jois; Naveen Chilamkurti; et al. ExoCarta: A Web-Based Compendium of Exosomal Cargo.. Journal of Molecular Biology 2015, 428, 688-692, 10.1016/j.jmb.2015.09.019.
- Hina Kalra; Richard J Simpson; Hong Ji; Elena Aikawa; Peter Altevogt; Philip Askenase; Vincent C. Bond; Francesc E. Borràs; Xandra Breakefield; Vivian Budnik; et al. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation.. PLOS Biology 2012, 10, e1001450, 10.1371/journal.pbio.1001450.
- Dae-Kyum Kim; Jaewook Lee; Richard J Simpson; Jan Lötvall; Yong Song Gho; EVpedia: A community web resource for prokaryotic and eukaryotic extracellular vesicles research. Seminars in Cell & Developmental Biology 2015, 40, 4-7, 10.1016/j.semcdb.2015.02.005.
- M Record; Kevin Carayon; Marc Poirot; Sandrine Silvente-Poirot; Exosomes as new vesicular lipid transporters involved in cell–cell communication and various pathophysiologies. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids 2014, 1841, 108-120, 10.1016/j.bbalip.2013.10.004.
- Karine Laulagnier; David Grand; Arnaud Dujardin; Safouane Hamdi; Hélène Vincent-Schneider; Danielle Lankar; Jean-Pierre Salles; Christian Bonnerot; Bertrand Perret; M Record; et al. PLD2 is enriched on exosomes and its activity is correlated to the release of exosomes. FEBS Letters 2004, 572, 11-14, 10.1016/j.febslet.2004.06.082.
- Clotilde Théry; Kenneth W. Witwer; Elena Aikawa; Maria Jose Alcaraz; Johnathon D Anderson; Ramaroson Andriantsitohaina; Anna Antoniou; Tanina Arab; Fabienne Archer; Georgia K Atkin-Smith; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of Extracellular Vesicles 2018, 7, 1535750, 10.1080/20013078.2018.1535750.
- Nobuyoshi Kosaka; Haruhisa Iguchi; Keitaro Hagiwara; Yusuke Yoshioka; Fumitaka Takeshita; Takahiro Ochiya; Neutral Sphingomyelinase 2 (nSMase2)-dependent Exosomal Transfer of Angiogenic MicroRNAs Regulate Cancer Cell Metastasis. Journal of Biological Chemistry 2013, 288, 10849-10859, 10.1074/jbc.m112.446831.
- Carolina Villarroya-Beltri; Cristina Gutiérrez‐Vázquez; Fátima Sánchez-Cabo; Daniel Pérez-Hernandez; Jesús Vázquez; Noa Beatriz Martín-Cófreces; Dannys Jorge Martínez-Herrera; Alberto Pascual-Montano; María Mittelbrunn; Francisco Sánchez-Madrid; et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nature Communications 2013, 4, 2980, 10.1038/ncomms3980.
- Laura Santangelo; Giorgio Giurato; Carla Cicchini; C. Montaldo; Carmine Mancone; Roberta Tarallo; Cecilia Battistelli; Tonino Alonzi; Alessandro Weisz; Marco Tripodi; et al. The RNA-Binding Protein SYNCRIP Is a Component of the Hepatocyte Exosomal Machinery Controlling MicroRNA Sorting. Cell Reports 2016, 17, 799-808, 10.1016/j.celrep.2016.09.031.
- Matthew J Shurtleff; Morayma M Temoche-Diaz; Kate V Karfilis; Sayaka Ri; Randy Schekman; Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. eLife 2016, 5, 5003, 10.7554/eLife.19276.
- Danijela Koppers-Lalic; Michael Hackenberg; Irene V. Bijnsdorp; Monique A.J. Van Eijndhoven; Payman Sadek; Daud Sie; Nicoletta Zini; Jaap M. Middeldorp; Bauke Ylstra; Renee X. De Menezes; et al. Nontemplated Nucleotide Additions Distinguish the Small RNA Composition in Cells from Exosomes. Cell Reports 2014, 8, 1649-1658, 10.1016/j.celrep.2014.08.027.
- Jasenka Guduric-Fuchs; Anna O’Connor; Bailey Camp; Christina L O'neill; Reinhold J. Medina; David A. Simpson; Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genomics 2012, 13, 357-357, 10.1186/1471-2164-13-357.