HAUSP (herpes virus-associated ubiquitin-specific protease), also known as Ubiquitin Specific Protease 7, plays critical roles in cellular processes, such as chromatin biology and epigenetics, through the regulation of different signaling pathways. HAUSP is a main partner of the “Epigenetic Code Replication Machinery,” ECREM, a large protein complex that includes several epigenetic players, such as the ubiquitin-like containing plant homeodomain (PHD) and an interesting new gene (RING), finger domains 1 (UHRF1), as well as DNA methyltransferase 1 (DNMT1), histone deacetylase 1 (HDAC1), histone methyltransferase G9a, and histone acetyltransferase TIP60. Due to its deubiquitinase activity and its ability to team up through direct interactions with several epigenetic regulators, mainly UHRF1, DNMT1, TIP60, the histone lysine methyltransferase EZH2, and the lysine-specific histone demethylase LSD1, HAUSP positions itself at the top of the regulatory hierarchies involved in epigenetic silencing of tumor suppressor genes in cancer.
- HAUSP
- UHRF1
- epigenetic
- cancer
1. Introduction
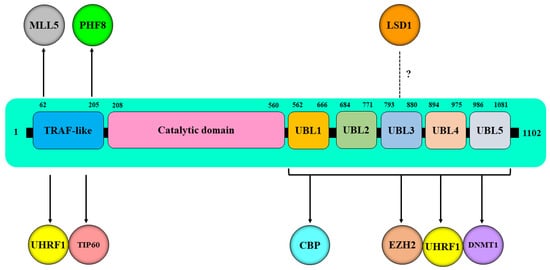
| Epi-Partner | Role of Epi-Partner | HAUSP Interaction Site | Epi-Partner Interaction Site | Related Epigenetic Events | Refs. |
|---|---|---|---|---|---|
| UHRF1 | Reader of both epigenetic marks (DNA methylation and histone code) | UBL1 domain (residues 560–664) |
A linker region encompassing amino acids 600–687 between the SRA and RING finger domains of UHRF1 | Promoting the stability of UHRF1 through HAUSP-dependent deubiquitination | [48] |
| TRAF-like domain | SRA domain | Promoting the stability of UHRF1 through HAUSP-dependent deubiquitination | [38] | ||
| (UBL1-2) domains | A polybasic region (PBR) in the C terminus | Promoting the association of the TTD-PHD domains of UHRF1 with chromatin and, hence, efficient H3K9me3 binding | [57] | ||
| DNMT1 | DNA methyltransferase 1 | C-terminal domain | Targeting sequence (TS) domain | Promoting the stability of DNMT1 through HAUSP-dependent deubiquitination | [38] |
| UBL domains (residues 560–1102) | KG linker (residues 1109–1119) | Promoting the stability of DNMT1 through acetylation of KG linker of DNMT1 | [49] | ||
| TIP60 | Histone acetyltransferase | TRAF-like domain | Increased levels of H2AK 5 and H4K5 | [50] | |
| CBP | Histone acetyltransferase | Region encompassing amino acids 600–687 | CH3 domain | Increased levels of H3K56Ac | [51] |
| MLL5 | Histone lysine methyltransferase | TRAF domain | Multiple domains | Increased levels of H3K4m3 | [53] |
| EZH2 | Histone lysine N-methyltransferase | C-terminal region | 489PRKKKRK495 region | Increased levels of H3K27m3 | [52] |
| LSD1 | Lysine specific demethylase 1 | [54] | |||
| A region encompassing amino acids 600–687 | Demethylation of H3K4me2 and H3K9me2 | [55] | |||
| PHF8 | Histone lysine demethylase | TRAF-like domain | The C-terminal region | Demethylation of H3K9me1,2, H3K27me2, and H4K20me1 | [56] |
2. Role of HAUSP in the Maintenance of DNA Methylation
2.1. HAUSP-Dependent Regulation of DNMT1 by Ubiquitination
The deubiquitinase activity of HAUSP regulates the stability and enzymatic activity of DNMT1 through several mechanisms. HAUSP can bind to DNMT1 and stabilize it through HAUSP-mediated deubiquitination [63]. High expression levels of DNMT1 protein have been found in human colon cancers, and this overexpression was correlated with HAUSP protein expression. Interestingly, the depletion of HAUSP in both human embryonic kidney cells and colorectal cancer cells resulted in an increase in DNMT1 ubiquitination and a reduction in DNMT1 protein expression. A similar increase in DNMT1 ubiquitination levels and a decrease in its protein expression were reported when HAUSP was knocked out in colorectal cancer cells [63]. By contrast, HAUSP overexpression in HAUSP knockout cells led to deubiquitination of DNMT1 and restoration of DNMT1 protein expression, suggesting that the deubiquitinase activity of HAUSP protects DNMT1 from proteasomal degradation, thereby promoting the stability and enzymatic activity of DNMT1 and the maintenance of DNA methylation [63].
One of the well-documented epigenetic regulators of the DNA methylation maintenance machinery is the epigenetic reader UHRF1, which has multiple functional domains [65,66,67]. Through its SRA domain, UHRF1 recognizes and binds hemi-methylated CpG islands, and via the same SRA domain, UHRF1 recruits DNMT1 to its correct position on chromatin to ensure faithful transmission of DNA methylation patterns during DNA replication [68,69,70].
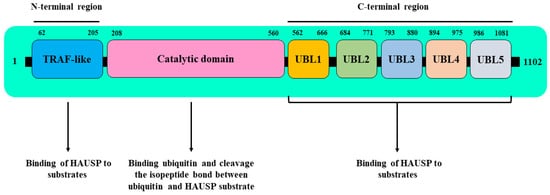
HAUSP binds to UHRF1 by its TRAF domain, but it also binds via its UBL domain (residues 560–664) to a linker region encompassing amino acids 600–687 between the SRA and RING finger domains of UHRF1, and this direct interaction is required for UHRF1 stability [48]. The downregulation of HAUSP decreased the expression levels of UHRF1 protein, whereas the overexpression of wild-type HAUSP, but not the catalytically inactive mutant HAUSP (C223S), significantly decreased the ubiquitination of UHRF1 and promoted its stability [48].
Obtaining a deep insight into how HAUSP deubiquitinase activity is involved in the stabilization of UHRF1 protein will help in understanding the regulatory role of HAUSP in the UHRF1-dependent maintenance of DNA methylation. In this regard, HAUSP was shown to regulate the stability of UHRF1 protein by targeting the ubiquitin ligase activity of the UHRF1 RING domain [38,65,81], which is used by UHRF1 to ubiquitinate itself (autoubiquitination) [65,66] or to ubiquitinate other substrates, mainly histone 3 [81,82,83]. Indeed, HAUSP, via its deubiquitinase activity, was shown to interfere with the E3 ubiquitin ligase activity of the RING domain of UHRF1, thereby eliminating the autoubiquitination of UHRF1 via the removal of ubiquitin adducts [38]. This led to the stabilization of UHRF1, indicating that HAUSP regulates the maintenance of DNA methylation through a direct association with UHRF1 [38].
2.2. HAUSP-Dependent Regulation of DNMT1 by Acetylation
3. Role of HAUSP in the Regulation of Histone Post-Translational Modifications
3.1. Role of HAUSP in the Regulation of Histone Monoubiquitination
3.2. Role of HAUSP in the Regulation of Histone Acetylation
3.3. Role of HAUSP in the Regulation of Histone Methylation
3.4. Role of HAUSP in the UHRF1-Mediated Readout of Histone H3 Lysine 9 Methylation
4. HAUSP Inhibitors as Promising Anticancer Agents
This entry is adapted from the peer-reviewed paper 10.3390/genes13010042
