The anti-virulence strategy is designed to prevent bacterial virulence factors produced by pathogenic bacteria from initiating and sustaining an infection.
- mono-ADP-ribosyltransferase toxins
- bacterial toxins
- protein crystallography
- anti-virulence strategy
1. Introduction

2. Anti-Virulence Approach
2.1. Virtual Screens
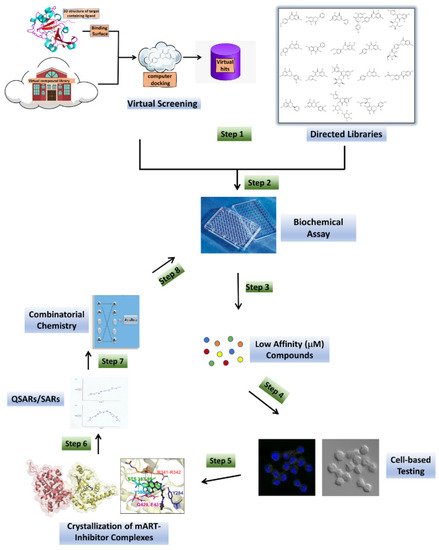
2.2. Directed Libraries
| Inhibitor | Chemical Name | Structure | a KD (μmol/L) |
b IC50 (μmol/L) |
|---|---|---|---|---|
| Acacetin | 5,7-dihydroxy-2-(4-methoxy phenyl)-4H-chromen-4-one | 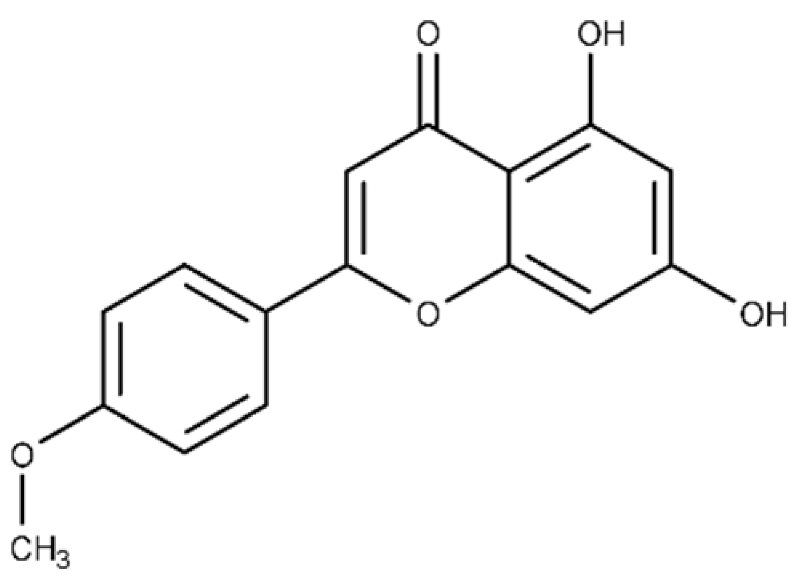 |
c ND | 28.0 ± 2.3 |
| Baicalein | 5,6,7-trihydroxy-2-phenyl-4H-chromen-4-one | 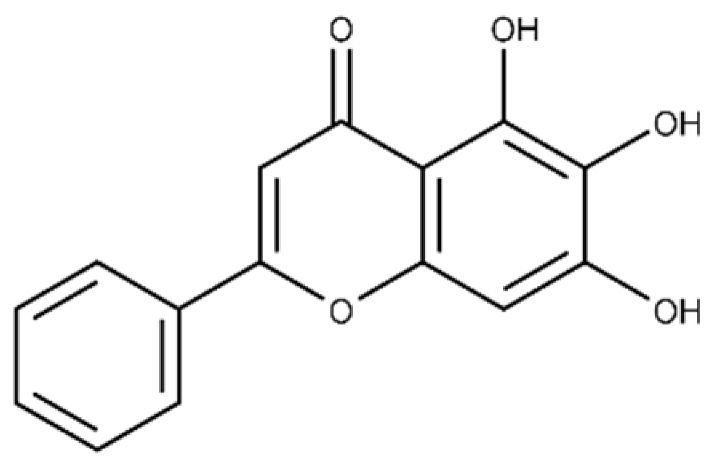 |
13.6 ± 1.5 | 10.7 ± 0.7 |
| Chrysin | 5,7-dihydroxy-2-phenyl-4H-chromen-4-one | 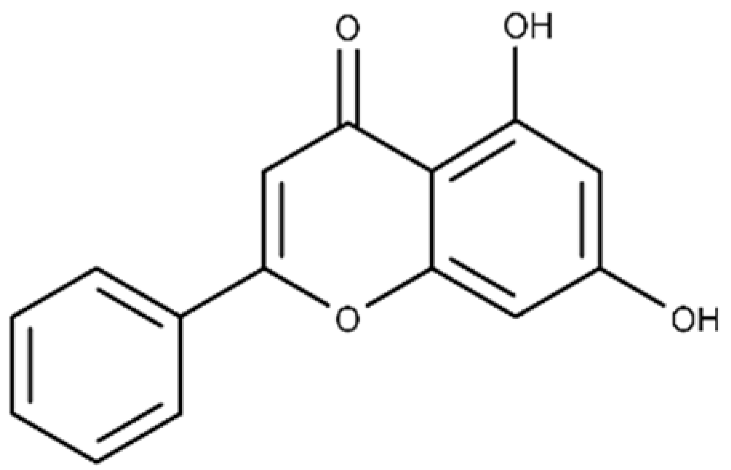 |
40.6 ± 4.7 | 49.2 ± 2.5 |
| Jaceosidin | 5,7-dihydroxy-2-(4-hydroxy-3-methoxyphenyl)-6-methoxy-4H-chromen-4-one | 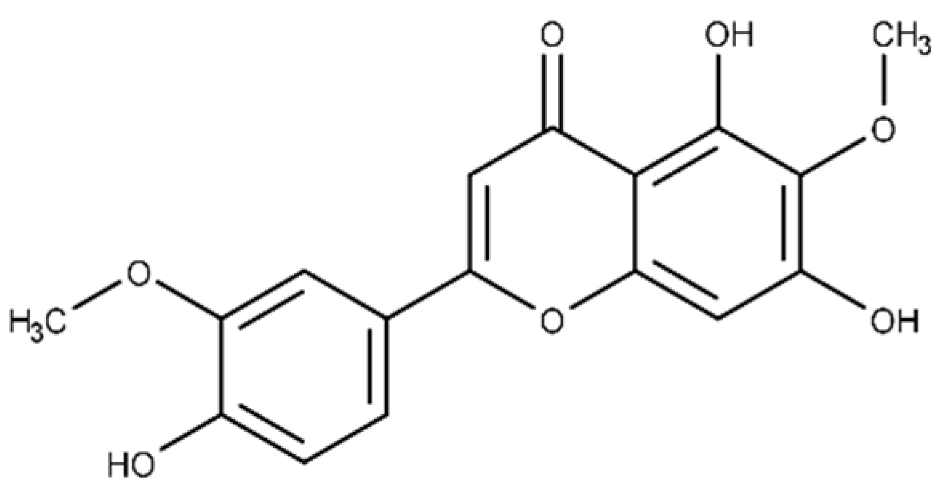 |
24.1 ± 2.9 | 73.9 ± 4.7 |
| Kaempferol | 3,5,7-trihydroxy-2-(4-hydroxyphenyl)-4H-chromen-4-one | 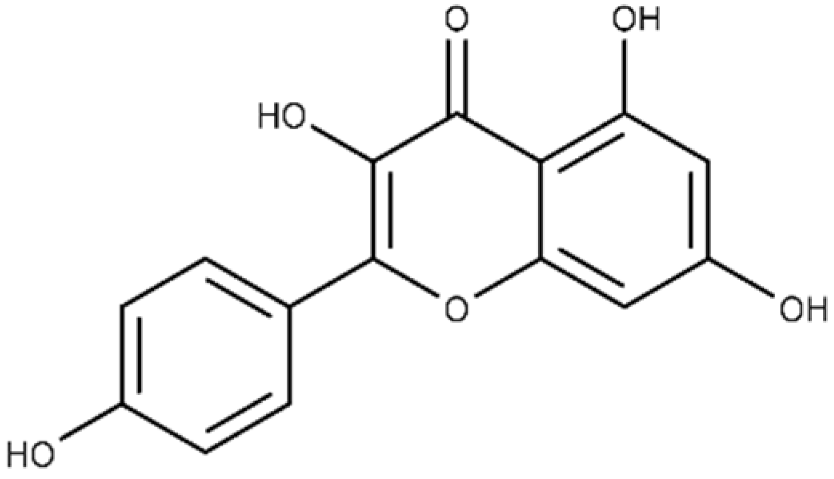 |
37.3 ± 3.6 | 79.9 ± 6.0 |
| Luteolin | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-chromen-4-one | 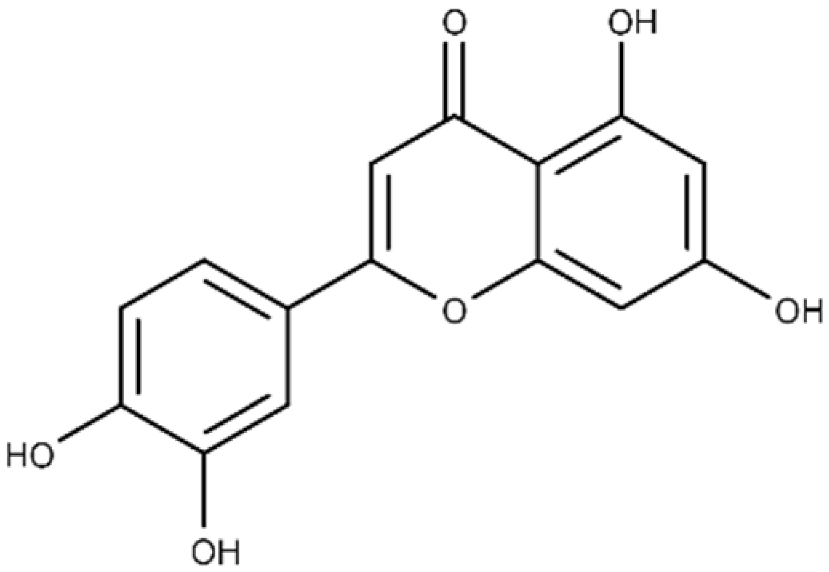 |
24.0 ± 1.5 | 68.1 ± 1.8 |
| Morin | 2-(2,4-dihydroxyphenyl)-3,5,7-trihydroxy-4H-chromen-4-one | 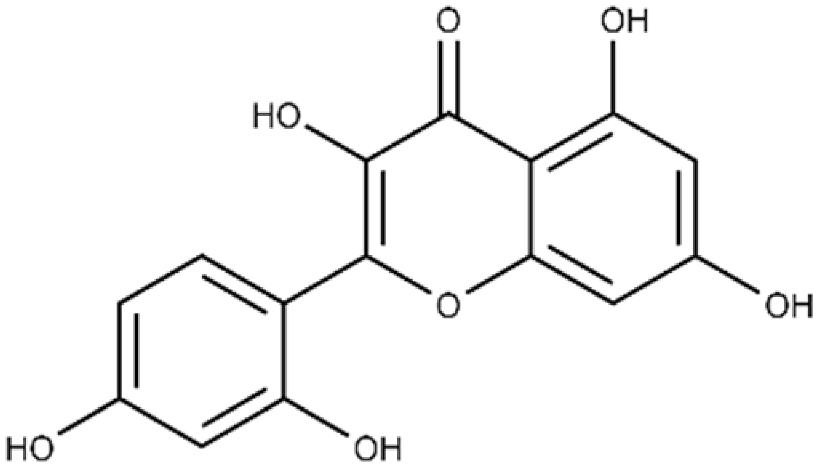 |
32.8 ± 11.7 | 109 ± 4.8 |
| Quercetin | 2-(3,4-dihydroxyphenyl)-3,5,7-trihydroxy-4H-chromen-4-one | 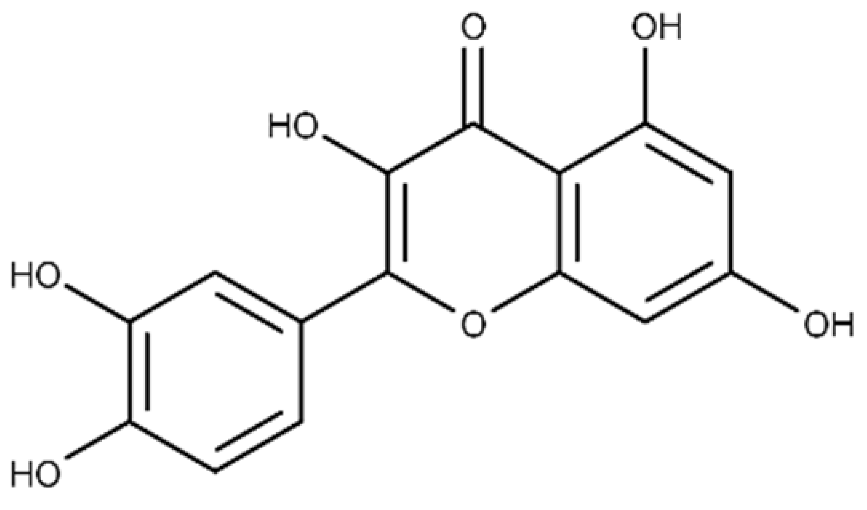 |
19.9 ± 0.4 | 24.3 ± 0.8 |
| M3 | N-{[(3R)-1-{1H-pyrazolo[3,4-d]pyrimidin-4- yl}piperidin-3-yl]methyl}methanesulfon amide |
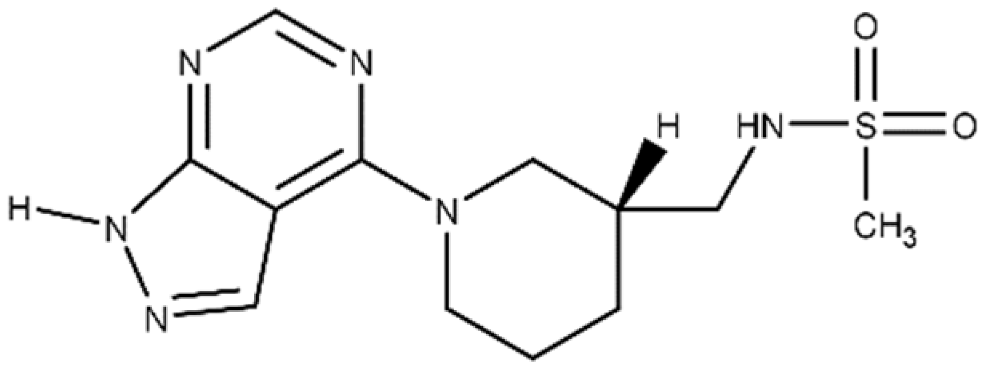 |
a The binding affinity of inhibitors to Plx2A (KD) were calculated from the binding isotherms generated from the quenching of intrinsic Trp fluorescence of the protein upon titration with the inhibitor (ligand). The values are the mean ± S.D. from three separate experiments. b The IC50 values were calculated from the fit to a dose–response curve of Plx2A GH activity against the concentration of inhibitor/flavonoid compounds to produce a directed library of 20 compounds for experimental testing against Plx2A activity [9]. c ND, not determined.
2.3. Biochemical Assay
2.4. Cell-Based Testing
2.5. Crystallization of mART-Inhibitor Complexes
2.6. SARs/QSARs and Combinatorial Chemistry
3. Anti-Virulence Agents against American Foulbrood
3.1. Paenibacillus Larvae
3.2. C3larvin Toxin
This entry is adapted from the peer-reviewed paper 10.3390/microorganisms9122514
References
- Jian, Z.; Zeng, L.; Xu, T.; Sun, S.; Yan, S.; Yang, L.; Huang, Y.; Jia, J.; Dou, T. Antibiotic resistance genes in bacteria: Occurrence, spread, and control. J. Basic Microbiol. 2021, 62, 1049–1070.
- Rubey, K.M.; Brenner, J.S. Nanomedicine to fight infectious disease. Adv. Drug Deliv. Rev. 2021, 179, 113996.
- Tompson, A.C.; Manderson, L.; Chandler, C.I.R. Understanding antibiotic use: Practices, structures and networks. JAC Antimicrob. Resist. 2021, 3, dlab150.
- Mousavi, S.M.; Babakhani, S.; Moradi, L.; Karami, S.; Shahbandeh, M.; Mirshekar, M.; Mohebi, S.; Moghadam, M.T. Bacteriophage as a Novel Therapeutic Weapon for Killing Colistin-Resistant Multi-Drug-Resistant and Extensively Drug-Resistant Gram-Negative Bacteria. Curr. Microbiol. 2021, 78, 4023–4036.
- Bhardwaj, S.; Bhatia, S.; Singh, S.; Franco, F., Jr. Growing emergence of drug-resistant Pseudomonas aeruginosa and attenuation of its virulence using quorum sensing inhibitors: A critical review. Iran. J. Basic Med. Sci. 2021, 24, 699–719.
- Kang, D.; Zhang, L.; Kirienko, N.V. High-Throughput Approaches for the Identification of Pseudomonas aeruginosa Antivirulents. mBio 2021, 12, e02240-20.
- Cherier, D.; Patin, D.; Blanot, D.; Touze, T.; Barreteau, H. The Biology of Colicin M and Its Orthologs. Antibiotics 2021, 10, 1109.
- Hotinger, J.A.; Morris, S.T.; May, A.E. The Case against Antibiotics and for Anti-Virulence Therapeutics. Microorganisms 2021, 9, 2049.
- Ebeling, J.; Pieper, F.; Gobel, J.; Knispel, H.; McCarthy, M.; Goncalves, M.; Turner, M.; Merrill, A.R.; Genersch, E. Anti-Virulence Strategy against the Honey Bee Pathogenic Bacterium Paenibacillus larvae via Small Molecule Inhibitors of the Bacterial Toxin Plx2A. Toxins 2021, 13, 607.
- Ford, C.A.; Hurford, I.M.; Cassat, J.E. Antivirulence Strategies for the Treatment of Staphylococcus aureus Infections: A Mini Review. Front. Microbiol. 2020, 11, 632706.
- Lim, K.Y.L.; Mullally, C.A.; Haese, E.C.; Kibble, E.A.; McCluskey, N.R.; Mikucki, E.C.; Thai, V.C.; Stubbs, K.A.; Sarkar-Tyson, M.; Kahler, C.M. Anti-Virulence Therapeutic Approaches for Neisseria gonorrhoeae. Antibiotics 2021, 10, 103.
- Zhang, D.; Gan, R.Y.; Zhang, J.R.; Farha, A.K.; Li, H.B.; Zhu, F.; Wang, X.H.; Corke, H. Antivirulence properties and related mechanisms of spice essential oils: A comprehensive review. Compr. Rev. Food Sci. Food Saf. 2020, 19, 1018–1055.
- Stewart, D.; Anwar, F.; Vedantam, G. Anti-virulence strategies for Clostridioides difficile infection: Advances and roadblocks. Gut Microbes 2020, 12, 1802865.
- Yakovlieva, L.; Fulleborn, J.A.; Walvoort, M.T.C. Opportunities and Challenges of Bacterial Glycosylation for the Development of Novel Antibacterial Strategies. Front. Microbiol. 2021, 12, 745702.
- Lakemeyer, M.; Zhao, W.; Mandl, F.A.; Hammann, P.; Sieber, S.A. Thinking outside the box—Novel antibacterials to tackle the resistance crisis. Angew. Chem. Int. Ed. 2018, 57, 14440–14475.
- Lugo, M.R.; Merrill, A.R. Development of Anti-Virulence Therapeutics against Mono-ADP-Ribosyltransferase Toxins. Toxins 2020, 13, 16.
- Ahmad-Mansour, N.; Loubet, P.; Pouget, C.; Dunyach-Remy, C.; Sotto, A.; Lavigne, J.P.; Molle, V. Staphylococcus aureus Toxins: An Update on Their Pathogenic Properties and Potential Treatments. Toxins 2021, 13, 677.
- Hu, H.; Liu, M.; Sun, S. Pore-Forming Toxins During Bacterial Infection: Molecular Mechanisms and Potential Therapeutic Targets. Drug Des. Dev. Ther. 2021, 15, 3773–3781.
- Kim, B.S. Spatiotemporal Regulation of Vibrio Exotoxins by HlyU and Other Transcriptional Regulators. Toxins 2020, 12, 544.
- Vij, R.; Hube, B.; Brunke, S. Uncharted territories in the discovery of antifungal and antivirulence natural products from bacteria. Comput. Struct. Biotechnol. J. 2021, 19, 1244–1252.
- Hotinger, J.A.; Pendergrass, H.A.; May, A.E. Molecular Targets and Strategies for Inhibition of the Bacterial Type III Secretion System (T3SS); Inhibitors Directly Binding to T3SS Components. Biomolecules 2021, 11, 316.
- Collier, R.J. Understanding the mode of action of diphtheria toxin: A perspective on progress during the 20th century. Toxicon 2001, 39, 1793–1803.
- Lyons, B.; Ravulapalli, R.; Lanoue, J.; Lugo, M.R.; Dutta, D.; Carlin, S.; Merrill, A.R. Scabin, a Novel DNA-acting ADP-ribosyltransferase from Streptomyces scabies. J. Biol. Chem. 2016, 291, 11198–11215.
- Simon, N.C.; Aktories, K.; Barbieri, J.T. Novel bacterial ADP-ribosylating toxins: Structure and function. Nat. Rev. Microbiol. 2014, 12, 599–611.
- Grimaldi, G.; Corda, D.; Catara, G. From toxins to mammalian enzymes: The diversity of mono-ADP-ribosylation. Front. Biosci. Landmark Ed. 2015, 20, 389–404.
- Yoshida, T.; Tsuge, H. Common Mechanism for Target Specificity of Protein- and DNA-Targeting ADP-Ribosyltransferases. Toxins 2021, 13, 40.
- Aktories, K.; Lang, A.E.; Schwan, C.; Mannherz, H.G. Actin as target for modification by bacterial protein toxins. FEBS J. 2011, 278, 4526–4543.
- Vogelsgesang, M.; Pautsch, A.; Aktories, K. C3 exoenzymes, novel insights into structure and action of Rho-ADP-ribosylating toxins. Naunyn Schmiedeberg’s Arch. Pharmacol. 2007, 374, 347–360.
- Fieldhouse, R.J.; Turgeon, Z.; White, D.; Merrill, A.R. Cholera- and anthrax-like toxins are among several new ADP-ribosyltransferases. PLoS Comput. Biol. 2010, 6, e1001029.
- Fieldhouse, R.J.; Merrill, A.R. Needle in the haystack: Structure-based toxin discovery. Trends Biochem. Sci. 2008, 33, 546–556.
- Tremblay, O.; Thow, Z.; Geddes-McAlister, J.; Merrill, A.R. Several New Putative Bacterial ADP-Ribosyltransferase Toxins Are Revealed from In Silico Data Mining, Including the Novel Toxin Vorin, Encoded by the Fire Blight Pathogen, Erwinia amylovora. Toxins 2020, 12, 792.
- Remmert, M.; Biegert, A.; Hauser, A.; Soding, J. HHblits: Lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat. Methods 2011, 9, 173–175.
- Soding, J.; Remmert, M.; Biegert, A.; Lupas, A.N. HHsenser: Exhaustive transitive profile search using HMM-HMM comparison. Nucleic Acids Res. 2006, 34, W374–W378.
- Turgeon, Z.; White, D.; Jorgensen, R.; Visschedyk, D.; Fieldhouse, R.J.; Mangroo, D.; Merrill, A.R. Yeast as a tool for characterizing mono-ADP-ribosyltransferase toxins. FEMS Microbiol. Lett. 2009, 300, 97–106.
- Swann, S.L.; Brown, S.P.; Muchmore, S.W.; Patel, H.; Merta, P.; Locklear, J.; Hajduk, P.J. A unified, probabilistic framework for structure- and ligand-based virtual screening. J. Med. Chem. 2011, 54, 1223–1232.
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749.
- Schneider, G. Virtual screening: An endless staircase? Nat. Rev. Drug Discov. 2010, 9, 273–276.
- Rollinger, J.M.; Stuppner, H.; Langer, T. Virtual screening for the discovery of bioactive natural products. Prog. Drug Res. 2008, 65, 211, 213–249.
- Zhu, T.; Cao, S.; Su, P.C.; Patel, R.; Shah, D.; Chokshi, H.B.; Szukala, R.; Johnson, M.E.; Hevener, K.E. Hit identification and optimization in virtual screening: Practical recommendations based on a critical literature analysis. J. Med. Chem. 2013, 56, 6560–6572.
- Jorgensen, W.L. Efficient drug lead discovery and optimization. Acc. Chem. Res. 2009, 42, 724–733.
- Cerqueira, N.M.; Sousa, S.F.; Fernandes, P.A.; Ramos, M.J. Virtual screening of compound libraries. Methods Mol. Biol. 2009, 572, 57–70.
- Sousa, S.F.; Cerqueira, N.M.; Fernandes, P.A.; Ramos, M.J. Virtual screening in drug design and development. Comb. Chem. High Throughput Screen. 2010, 13, 442–453.
- Irwin, J.J.; Raushel, F.M.; Shoichet, B.K. Virtual screening against metalloenzymes for inhibitors and substrates. Biochemistry 2005, 44, 12316–12328.
- Danishuddin, M.; Khan, A.U. Structure based virtual screening to discover putative drug candidates: Necessary considerations and successful case studies. Methods 2015, 71, 135–145.
- Elseginy, S.A. Virtual screening and structure-based 3D pharmacophore approach to identify small-molecule inhibitors of SARS-CoV-2 Mpro. J. Biomol. Struct. Dyn. 2021, 1–17.
- Shniffer, A.; Visschedyk, D.D.; Ravulapalli, R.; Suarez, G.; Turgeon, Z.J.; Petrie, A.A.; Chopra, A.K.; Merrill, A.R. Characterization of an actin-targeting ADP-ribosyltransferase from Aeromonas hydrophila. J. Biol. Chem. 2012, 287, 37030–37041.
- Ravulapalli, R.; Lugo, M.R.; Pfoh, R.; Visschedyk, D.; Poole, A.; Fieldhouse, R.J.; Pai, E.F.; Merrill, A.R. Characterization of Vis Toxin, a Novel ADP-Ribosyltransferase from Vibrio splendidus. Biochemistry 2015, 54, 5920–5936.
- Krska, D.; Ravulapalli, R.; Fieldhouse, R.J.; Lugo, M.R.; Merrill, A.R. C3larvin Toxin, an ADP-ribosyltransferase from Paenibacillus larvae. J. Biol. Chem. 2015, 290, 1639–1653.
- Panche, A.N.; Diwan, A.D.; Chandra, S.R. Flavonoids: An overview. J. Nutr. Sci. 2016, 5, e47.
- Babii, C.; Mihalache, G.; Bahrin, L.G.; Neagu, A.N.; Gostin, I.; Mihai, C.T.; Sarbu, L.G.; Birsa, L.M.; Stefan, M. A novel synthetic flavonoid with potent antibacterial properties: In vitro activity and proposed mode of action. PLoS ONE 2018, 13, e0194898.
- Cushnie, T.P.T.; Lamb, A.J. Antimicrobial activity of flavonoids. Int. J. Antimicrob. Agents 2005, 26, 343–356.
- Farhadi, F.; Khameneh, B.; Iranshahi, M.; Iranshahy, M. Antibacterial activity of flavonoids and their structure-activity relationship: An update review. Phytother. Res. 2019, 33, 13–40.
- Goel, S.; Khulbe, M.; Aggarwal, A.; Kathuria, A. Recent advances in continuous flow synthesis of heterocycles. Mol. Divers. 2021.
- Egami, H.; Hamashima, Y. Practical and Scalable Organic Reactions with Flow Microwave Apparatus. Chem. Rec. 2019, 19, 157–171.
- Morin, M.A.; Zhang, W.P.; Mallik, D.; Organ, M.G. Sampling and Analysis in Flow: The Keys to Smarter, More Controllable, and Sustainable Fine-Chemical Manufacturing. Angew. Chem. Int. Ed. 2021, 60, 20606–20626.
- Lipinski, C.A. Rule of five in 2015 and beyond: Target and ligand structural limitations, ligand chemistry structure and drug discovery project decisions. Adv. Drug Deliv. Rev. 2016, 101, 34–41.
- Khalifa, S.A.M.; Elshafiey, E.H.; Shetaia, A.A.; El-Wahed, A.A.A.; Algethami, A.F.; Musharraf, S.G.; AlAjmi, M.F.; Zhao, C.; Masry, S.H.D.; Abdel-Daim, M.M.; et al. Overview of Bee Pollination and Its Economic Value for Crop Production. Insects 2021, 12, 688.
- Poppinga, L.; Genersch, E. Molecular pathogenesis of American Foulbrood: How Paenibacillus larvae kills honey bee larvae. Curr. Opin. Insect Sci. 2015, 10, 29–36.
- Applegate, J.R., Jr.; Petritz, O.A. Common and Emerging Infectious Diseases of Honeybees (Apis mellifera). Vet. Clin. Exot. Anim. Pract. 2020, 23, 285–297.
- Ebeling, J.; Knispel, H.; Hertlein, G.; Funfhaus, A.; Genersch, E. Biology of Paenibacillus larvae, a deadly pathogen of honey bee larvae. Appl. Microbiol. Biotechnol. 2016, 100, 7387–7395.
- Genersch, E. American Foulbrood in honeybees and its causative agent, Paenibacillus larvae. J. Invertebr. Pathol. 2010, 103 (Suppl. 1), S10–S19.
- Beims, H.; Bunk, B.; Erler, S.; Mohr, K.I.; Sproer, C.; Pradella, S.; Gunther, G.; Rohde, M.; von der Ohe, W.; Steinert, M. Discovery of Paenibacillus larvae ERIC V: Phenotypic and genomic comparison to genotypes ERIC I–IV reveal different inventories of virulence factors which correlate with epidemiological prevalences of American Foulbrood. Int. J. Med. Microbiol. 2020, 310, 151394.
- Djukic, M.; Brzuszkiewicz, E.; Funfhaus, A.; Voss, J.; Gollnow, K.; Poppinga, L.; Liesegang, H.; Garcia-Gonzalez, E.; Genersch, E.; Daniel, R. How to kill the honey bee larva: Genomic potential and virulence mechanisms of Paenibacillus larvae. PLoS ONE 2014, 9, e90914.
- Funfhaus, A.; Poppinga, L.; Genersch, E. Identification and characterization of two novel toxins expressed by the lethal honey bee pathogen Paenibacillus larvae, the causative agent of American foulbrood. Environ. Microbiol. 2013, 15, 2951–2965.
- Ebeling, J.; Funfhaus, A.; Knispel, H.; Krska, D.; Ravulapalli, R.; Heney, K.A.; Lugo, M.R.; Merrill, A.R.; Genersch, E. Characterization of the toxin Plx2A, a RhoA-targeting ADP-ribosyltransferase produced by the honey bee pathogen Paenibacillus larvae. Environ. Microbiol. 2017, 19, 5100–5116.
- Ebeling, J.; Funfhaus, A.; Genersch, E. The Buzz about ADP-Ribosylation Toxins from Paenibacillus larvae, the Causative Agent of American Foulbrood in Honey Bees. Toxins 2021, 13, 151.
- Turner, M.; Tremblay, O.; Heney, K.A.; Lugo, M.R.; Ebeling, J.; Genersch, E.; Merrill, A.R. Characterization of C3larvinA, a novel RhoA-targeting ADP-ribosyltransferase toxin produced by the honey bee pathogen, Paenibacillus larvae. Biosci. Rep. 2020, 40, BSR20193405.
