Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Anatomy & Morphology
CD44, a non-kinase cell surface transmembrane glycoprotein, has been widely implicated as a cancer stem cell (CSC) marker in several cancers. Cells overexpressing CD44 possess several CSC traits, such as self-renewal and epithelial-mesenchymal transition (EMT) capability, as well as a resistance to chemo- and radiotherapy.
- CD44
- regulation
- tumourigenesis
1. CD44 Structure and Isoforms
The full-length CD44 gene comprises 20 exons, with the constant exons 1–5 and 16–20 encoding the N-terminal and C-terminal domains respectively, which are homologous domains shared by all CD44 family members [10]. The smallest and the most expressed CD44 isoform is the CD44 standard (CD44s), constructed of ten constant exons with no variant exons [11]. The other isoform, the CD44 variant (CD44v), differs from CD44s by the insertion or excision of alternatively spliced exons between the N-terminal and C-terminal domains [12]. Tolg et al. [13] confirmed that besides ten constant exons, the mouse and rat genome has at least ten variant exons, all of which can be combined alternatively into CD44 mRNA. They suggested that the variant exons be numbered by the exon code v1 to v10. Screaton et al. [14] described the structure of the human CD44 gene, reporting that it contains 19 exons crossing some 50 kilobases of DNA with ten constant exons and nine variant exons coded v2–v10 [15,16]. CD44v isoforms may contain a single variant exon such as CD44v3 or CD44v6, or a combination of variant exons such as CD44v3–v7 and CD44v8-v10. Individual cells can continually alter the splicing of CD44 pre-mRNA, resulting in many possible combinations of these variant exons, giving the potential for great diversity [12].
The CD44 protein has four primary characteristic regions: the extracellular region, the stem region (standard stem region and/or variable stem region), the transmembrane region (TM), and the short C-terminal intracellular/cytoplasmic (CP) region [17]. The extracellular part consists of seven extracellular domains (1–5, 6 and 7 of the constant exons) including N-terminal domains (ligand-binding region). The stem region (alternative splicing area) has an insertion of one or more of the variant exons between exon 5 and exon 6. The transmembrane region is encoded by a single exon (exon 8), whereas the cytoplasmic region is encoded by exon 10 or exon 9. However, exon 9 is spliced out in almost all CD44 cDNA isoforms [12,18]. Several isoforms of the human CD44 molecule are associated with tumour progression and stemness in various cancers, such as breast cancer [19], gliomas [20,21], head and neck squamous cell carcinoma [22], pancreatic cancer [23,24], prostate cancer [25] and colorectal cancer [26,27] (Figure 1 and Table 1). The complexity of the CD44 protein is further augmented by post-translational modifications including variance glycosylation with O-glycans, N-glycans and glycosaminoglycans, such as chondroitin sulphate and heparan sulphate [17]. Due to these side-chain attachments, the conserved format of CD44 (37 kDa) is enlarged to 80–100 kDa with some isoforms surpassing 200 kDa due to a high level of glycosylation [12]. An illustration of CD44 protein structure is shown in Figure 2.
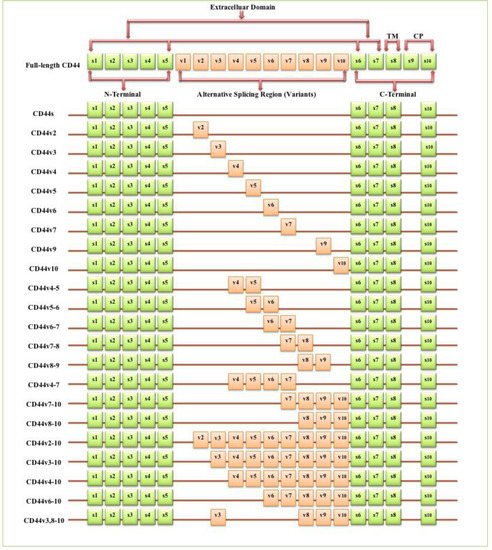
Figure 1. Schematic diagram of the mouse CD44 gene and most CD44 isoforms involved in cancer progression. The full-length CD44 gene contains 20 exons in mice and 19 exons in humans, with the constant exons 1–5 and 16–20 encoding the N-terminal and the C-terminal domains. CD44 standard (CD44s) is encoded by these ten constant exons and contains no variant exons, whereas the CD44 variant (CD44v) is produced by the alternative splicing of a variable insertion of nine extra exons in humans or ten extra exons in mice. These extra exons are exons 6-15, typically identified as (v1 to v10) in mice and the exons 7-15 identified as (v2 to v10) in humans and are located between the N-terminal and C-terminal domains. CD44v can contain one or multiple variant exons and exon 19 is spliced out in all CD44 isoforms. Abbreviations: CD44s, CD44 standard; CD44v, CD44 variant; s, standard; v, variant; TM, transmembrane; CP, cytoplasmic. Green boxes refer to the constant/standard exons. Orange boxes refer to the variant exons.
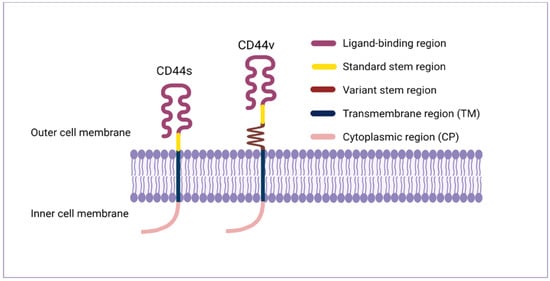
Figure 2. CD44 protein structure. The CD44 protein has four primary regions: the extracellular region consists of seven extracellular domains including N-terminal domains (ligand-binding region), the stem region (variable stem region and/or standard stem region) which is the alternative splicing area containing an insertion of one or more variant exons, the transmembrane region (TM), and the C-terminal cytoplasmic (CP) region.
Table 1. CD44 isoforms relevant to cancer progression. Abbreviations: CSCs, cancer stem cells; EMT, epithelial–mesenchymal transition; DFS, disease-free survival; OS, overall survival; TNM stage, tumour (T), node (N), and metastasis (M) stage; FIGO stage, the international federation of gynaecology and obstetrics stage; NHL, Non-Hodgkin’s lymphoma; HPV, human papillomavirus; MAPK, mitogen-activated protein kinase.
| CD44 Isoform | Association in Cancer Progress | Cancer Type | Ref |
|---|---|---|---|
| CD44, non-specified | Tumour cell aggregation, metastasis | Breast cancer | [19] |
| CD44, non-specified | Adhesion, migration, invasion | Glioblastoma | [20,21] |
| CD44, non-specified | Angiogenesis | Head and neck squamous carcinoma | [22] |
| CD44, non-specified | Invasion, metastasis, EMT, cancer progression, poor prognosis | Pancreatic cancer | [23,24] |
| CD44, non-specified | Proliferation, migration, invasion | Prostate Cancer | [25] |
| CD44, non-specified | Metastasis, poor differentiation, invasion | Colorectal cancer | [26,27] |
| CD44s | Tumour initiation, CSCs traits induction | Breast cancer | [28] |
| CD44s | Metastasis | Breast cancer | [29] |
| CD44s | EMT regulation, cancer progression | Breast cancer | [30] |
| CD44s | Poor DFS, poor OS, invasion, EMT | Hepatocellular carcinoma | [31] |
| CD44s | Invasion, metastasis, EMT, poor differentiation, chemotaxis |
Gallbladder cancer | [32] |
| CD44s | Proliferation, invasion, migration, EMT, stemness | Prostate cancer | [33] |
| CD44s | EMT, invasion, metastasis, chemoresistance | Pancreatic ductal adenocarcinoma | [34] |
| CD44s | EMT, radio-resistance | Pancreatic cancer | [35] |
| CD44v2 | Poor OS, advanced cancer stage | Colorectal cancer | [36] |
| CD44v2 | Poor OS, invasion | Pancreatic cancer | [37] |
| CD44v3 | Poor OS, invasion, metastasis | Oral squamous carcinoma | [38] |
| CD44v3 | Stem cells self-renewal | Myeloid leukaemia | [39] |
| CD44v3 | Metastasis | Colorectal adenocarcinoma | [40] |
| CD44v4 | Proliferation, migration, radio-resistance | Head and neck squamous carcinoma |
[41] |
| CD44v5 | High histological grade, poor differentiation, poor OS |
Hepatocellular carcinoma | [42] |
| CD44v6 | Tumour budding, invasion, metastasis | Oral squamous carcinoma | [43] |
| CD44v6 | Proliferation, invasion, adhesion, metastasis, EMT, chemo/radio-resistance |
Prostate cancer | [44] |
| CD44v6 | Local recurrence, invasion, metastasis | Tongue squamous carcinoma | [45] |
| CD44v6 | Tumour budding, locoregional failure (metastasis, local recurrence) | Colorectal cancer | [46] |
| CD44v6 | Proliferation, migration, radio-resistance | Head and neck squamous carcinoma |
[41] |
| CD44v6 | Metastasis | Colorectal adenocarcinoma | [40] |
| CD44v6 | Poor OS, invasion | Pancreatic cancer | [37] |
| CD44v6 | High histological grade, poor differentiation, poor OS |
Hepatocellular carcinoma | [42] |
| CD44v6 | Invasion, metastasis, poor OS, TNM stage | Pancreatic cancer | [47] |
| CD44v6 | FIGO stage, poor prognosis | Cervical cancer | [48] |
| CD44v6 | Metastasis, self-adhesion of aggressive NHL cells | Non-Hodgkin’s lymphoma | [49] |
| CD44v6 | Infiltration, metastasis | Oesophageal squamous carcinoma | [50] |
| CD44v6 | Proliferation, myofibroblastic differentiation | Gastric cancer | [51] |
| CD44v7 | Proliferation, migration, radio-resistance | Head and neck squamous carcinoma |
[41] |
| CD44v9 | Increased tumourigenicity | Gallbladder cancer | [32] |
| CD44v9 | Invasion, metastasis, poor OS, TNM stage | Pancreatic cancer | [47] |
| CD44v9 | Proliferation, invasion, migration, EMT | Cholangiocarcinoma | [52] |
| CD44v9 | Invasion, migration, worse prognosis | Bladder cancer | [53] |
| CD44v10 | High histological grade, poor differentiation, poor OS |
Hepatocellular carcinoma | [42] |
| CD44v10 | Histological grade, clinical and pathological stage, poor survival |
Renal carcinoma | [54] |
| CD44v10 | Migration, metastasis, promote tumourigenesis |
Breast cancer | [55,56] |
| CD44v4-5 | Infiltration, metastasis | Oesophageal squamous carcinoma | [50] |
| CD44v4-5 | Poor differentiation | Non-small cell lung carcinoma | [57] |
| CD44v5-6 | Proliferation, KRAS/MAPK signalling, promoting tumour development | Lung adenocarcinoma | [58] |
| CD44v6-7 | Metastasis | Pancreatic adenocarcinoma | [11] |
| CD44v7-8 | High histological grade, poor differentiation, poor OS |
Hepatocellular carcinoma | [42] |
| CD44v7-8 | FIGO stage, poor prognosis | Cervical cancer | [48] |
| CD44v7-8 | Invasion, high-risk HPV infection | Uterine cervical squamous carcinoma |
[59] |
| CD44v8-9 | Proliferation, KRAS/MAPK signalling, promoting tumour development | Lung adenocarcinoma | [58] |
| CD44v4-7 | Metastasis | Pancreatic adenocarcinoma | [11] |
| CD44v7-10 | Invasion | Prostate cancer | [60] |
| CD44v8-10 | Migration, metastasis, sphere formation | Breast cancer | [61] |
| CD44v8-10 | Tumour initiation, CSCs traits induction | Gastric cancer | [62] |
| CD44v8-10 | Metastasis | Lung cancer | [63] |
| CD44v8-10 | Metastasis, relapse | Gastric cancer | [64] |
| CD44v8-10 | Poor prognosis, chemo/radio-resistance | Oesophageal squamous carcinoma | [65] |
| CD44v8-10 | Chemoresistance | Urothelial cancer | [66] |
| CD44v2-10 | CSCs traits induction, tumour subtype, oncogenic signalling pathways | Breast cancer | [67] |
| CD44v3-10 | CSCs traits induction, tumour subtype, oncogenic signalling pathways | Breast cancer | [67] |
| CD44v3-10 | Metastasis, self-adhesion of aggressive NHL cells | Non-Hodgkin’s lymphoma | [49] |
| CD44v4-10 | Tumour initiation, wild-type phenotype | Intestinal cancer | [15] |
| CD44v6-10 | Metastasis, self-adhesion of aggressive NHL cells | Non-Hodgkin’s lymphoma | [49] |
| CD44v6-10 | Metastasis, relapse | Gastric cancer | [64] |
| CD44v3, 8-10 | Metastasis, relapse | Gastric cancer | [64] |
| CD44v3, 8-10 | Metastasis, migration | Breast cancer | [68] |
2. CD44 Expression in Normal Cells
CD44 is significantly expressed in lymphocytes, smooth muscle, fibroblasts and various types of epithelia and is involved in lymphocyte homing, cell adhesion and aggregation, cell migration, leukocyte activation, lymphopoiesis and myelopoiesis, angiogenesis and cytokine release [12,69]. CD44s was initially isolated from haematopoietic cells even though it is expressed in several other tissues including the liver, lung, pancreas, skin and central nervous system [12]. CD44s is expressed in adult tissues and embryo tissues from day 9.5 post coitum, whereas numerous isoforms of CD44v show a highly specialised expression pattern and are already in the egg cylinder at day 6.5 of development [70]. In contrast to CD44s, CD44v isoforms distribution is more restricted to a selected range of cells during specific stages of activation, maturation or development including macrophages, activated lymphocytes, keratinocytes and some epithelial cells such as in the stomach, bladder and uterine cervix [12] and many carcinomas. In normal tissues, CD44 isoforms play a role in hyaluronic acid (HA) metabolism regulation, whereby loss of CD44 expression disrupts HA metabolism and impairs hair regrowth, wound healing and keratinocyte proliferation [71].
3. CD44 Expression in Tumours
Numerous studies indicated that lymphoma, breast, colon and endometrial cancer have elevated levels of CD44 mRNA [69]. Increasing evidence also suggests that CD44 is extensively overexpressed in other cancer types including gallbladder, prostate, ovarian, oral squamous cell carcinoma and gastric cancer, correlating with aggressive biological behaviour and a poor prognosis [72]. The role of CD44 in tumours is not well defined, however, elevated levels of CD44 are associated with numerous malignant tumours. The physiological functions of CD44 indicate that it is involved in the metastasis of tumours [12]. For instance, lung adenocarcinoma cells show a high expression of CD44v, which correlates with enhanced CSCs characteristics, proliferation and resistance to chemotherapeutics [73], whereas these variants, especially CD44v6, are closely related to metastasis of pancreatic carcinoma cells [69]. Many studies have investigated CD44 expression levels in several cancers in comparison to their adjacent normal tissues and explored the relationship with tumour progression and clinicopathological outcomes by mining various publicly available databases, including The Cancer Genome Atlas (TCGA), Tumour Immune Estimation Resource (TIMER) database, Oncomine database, Gene Expression Profiling Interactive Analysis (GEPIA), In silico Transcriptomics (IST) database, R2 online database, SAGE Genie tools, and Human Gene Expression Map (HGEM) (Table 2 and Figure 3).
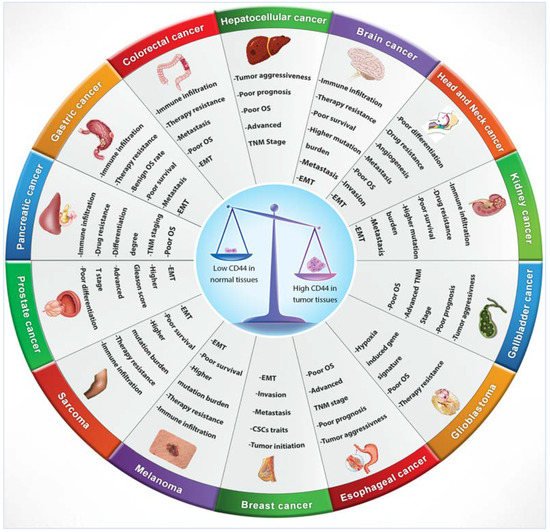
Figure 3. CD44 distribution in normal versus cancerous tissues and its correlation with clinical outcomes.
Table 2. Low and high CD44 expression in normal and tumour tissues respectively and association with clinical outcomes.
| Cancer Type | Correlation with Clinical Outcomes | Public Database | Reference |
|---|---|---|---|
| Gallbladder cancer, hepatocellular carcinoma, cholangiocarcinoma | Poor prognosis, advanced TNM stage, poor OS, aggressive tumour behaviour (proliferation, migration, invasion, clonogenicity) | TCGA database | [72] |
| Colon cancer, gastric cancer, brain cancer, stomach cancer, pancreatic cancer, liver cancer | Benign OS rate in gastric cancer, poor OS in colon cancer, TNM staging, differentiation degree, and poor survival in pancreatic cancer | SAGE Genie and Oncomine database | [74,75] |
| Head and neck squamous carcinoma | Poor OS, poor differentiation, angiogenesis, immune regulation, invasion | TCGA database | [76] |
| Head and neck squamous carcinoma | Pro-angiogenetic phenotype | TCGA database | [22] |
| Prostate cancer | Advanced T stage, higher Gleason score, poor differentiation | TCGA database | [77] |
| Colon adenocarcinoma | Therapy resistance | TCGA database and GEPIA | [78] |
| Head and neck squamous carcinoma, acute myeloid leukaemia (AML), lung carcinoma | Not specified | IST database and HGEM database | [79] |
| Glioblastoma | Poor OS, hypoxia-induced gene signature | TCGA database | [80] |
| Glioblastoma | Poor OS, therapy resistance | R2 online database | [81] |
| Invasive ductal breast carcinoma | Invasion, metastasis | TCGA database | [82] |
| Brain and CNS cancer, colorectal cancer, melanoma, sarcoma, gastric cancer, head and neck carcinoma, kidney cancer, oesophageal cancer, cholangiocarcinoma, pancreatic cancer | EMT, drug resistance, metastasis, immune infiltration and suppression features, poor survival, higher mutation burden, afflict older patients | Oncomine database and TIMER database | [83] |
This entry is adapted from the peer-reviewed paper 10.3390/biom11121850
This entry is offline, you can click here to edit this entry!
