Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Biochemistry & Molecular Biology
Transcription factors are key molecules in the regulation of gene expression in all organisms. The transcription factor LEAFY COTYLEDON 2 (LEC2), which belongs to the DNA-binding protein family, contains a B3 domain. The transcription factor is involved in the regulation of important plant biological processes such as embryogenesis, somatic embryo formation, seed storage protein synthesis, fatty acid metabolism, and other important biological processes.
- embryogenesis
- somatic embryogenesis
- plant growth
- seed development
- transcription factor
- LEC2
1. Introduction
Seeds are the key means by which terrestrial plants adapt to changing environments in the processes of evolution and diversification, and seed development occurs after zygotic embryogenesis. When seeds are in the maturation stage of growth and development, energy reserves continue to accumulate, while the seeds gain desiccation tolerance [1]. This process is strictly controlled at the transcription level, involving the AFL (ABI3/FUS3/LEC2) (ABA INSENSITIVE3/FUSCA3/LEAFY COTYLEDON 2) subfamily of B3 transcription factors (TFs). Researchers have shown that the B3 TF gene family developed within the green algae family 1200–725 million years ago, and the genes are present in all photosynthetic organisms [2]. AFL members interact with LEC1 (LEAFY COTYLEDON1) and LEC1-LIKE, which belong to the CCAAT-binding factors of the HAP3 family to control seed growth and development [3][4][5][6]. These genes have been collectively named L-AFL [7].
Members of the L-AFL family are considered to be the key regulatory TFs during the seed maturity stage [8]. The LEC2 TF establishes an ideal cellular environment for the formation of the zygotic embryo and its later stages of development [9][10]. Early embryonic development is the main period of expression of LEC2; at times, it is also expressed in vegetative organs [11][12]. LEC2 contains two domains named B2 and B3 [13]. It appears that the plant-specific B3 domain encoded by the LEC2 gene recognizes the conserved RY motif to transcriptionally regulate the expression of zygotic embryogenesis-specific genes and that it promotes somatic embryo formation at the maturation stage [12][14][15]. The LEC2 gene could be directly repressed by E2FA binding to an E2F-binding site during the seed maturation phase [16]. A ChIP assay suggested that PHABULOSA acts directly on the LEC2 promoter during embryogenesis [17]. An analysis using reporter genes indicated that LEC2 is negatively regulated by miRNA pathways during early embryogenesis [18]. miRNA is responsible, directly or indirectly, for repressing LEC2 in the embryo until it is required [17][18][19]. Retinoblastoma-related proteins facilitate seedling establishment by directly or indirectly repressing the promoters of late embryogenesis genes, including LEC2, during seed germination [20].
LEC2 participates in a variety of signaling pathways and regulates the expression of numerous crucial genes during the growth and development of plants. Early studies have shown that the mutation of LEC2 in Arabidopsis thaliana altered the morphology of the embryo and caused certain local defects in the seed protein stockpile and its desiccation tolerance [12][21]. In addition, LEC2 mutations halted the ability of somatic embryos to emerge from A. thaliana explants [22]. Through further in-depth exploration of the biological functions of LEC2, it was shown that the ectopic expression of LEC2 caused the accumulation of seed storage lipids and proteins in plant nutritive organs [23][24], further inducing vegetative cells to form somatic embryos without exogenous auxin or seed-specific genes being expressed in the leaves [12][23]. In A. thaliana, the function of LEC2 has been explained in terms of many aspects (Table 1).
Table 1. Biological function of LEC2 transcription factor in A. thaliana.
| Biological Function | Reference |
|---|---|
| Induces somatic embryos and embryonic development in vegetative cells | [4] |
| Initiates somatic embryo development | [12] |
| Regulates the expression of storage protein genes | [11] |
| Induces somatic embryogenesis | [22] |
| Triggers the stockpile of oil and seed-specific mRNAs | [23] |
| Induces maturation traits and auxin activity | [24] |
| Affects the contents of oil and protein, starch, and sucrose | [25] |
| Changes the shape and anatomy of leaves | [26] |
| Triggers the expression of genes encoding seed maturation and oil body protein regulators in trophic organization | [27] |
| Promotes embryogenic callus formation in roots | [28] |
| Controls the formation of lateral roots | [29] |
| Involved in early embryogenesis Participates in the development of somatic embryos |
[30] [31] |
2. The Effect of LEC2 on Plant Early Embryo Morphogenesis
In the process of plant early embryogenesis, specialized leaves called cotyledons are produced. Compared with ordinary leaves, these embryonic leaves have large differences in morphology and gene expression patterns. When AtLEC2 is mutated, the cotyledons undergo certain changes, including rounding in shape and the development of abnormal protrusions on the surface. Mutant cotyledons produce trichomes characteristic of leaves, indicating that AtLEC2 is important for maintaining cotyledon traits during early embryogenesis [32].
Recent research has shed new light on AtLEC2’s involvement in the development of early embryos. When plants undergo a long winter, the polycomb protein silences the potent flower repressor FLOWERING LOCUS C (FLC) that induces flowers to undergo vernalization. VIVIPAROUS1/ABI3-LIKE1 (VAL1) and VAL2 are necessary for this process [33][34]. LEC2 and FUS3 are also required for embryonic FLC reactivation in early embryos following parental vernalization [30]. Late flowering is dependent on FLC in non-vernalized plants. However, this phenomenon is suppressed in LEC2 and FUS3 seeds. Hence, LEC2 and FUS3 are involved in embryonic FLC reactivation. FLC reactivation is also suppressed by LEC2 in vernalized seedlings. In addition, the parental vernalization of T2 progeny from FUS3 plants caused a reduction in FLC expression in the seedlings of T3 progeny [30]. LEC2 and FUS3 bind to the cold memory element of FLC to reactivate FLC expression in early embryos following parental vernalization. The ectopic induction of LEC2 or FUS3 activity can antagonize FLC repression mediated by VAL1 and VAL2 in seedlings. The B3 domain TFs LEC2 and FUS3 can replace VAL1 and VAL2 to reverse the chromatin-mediated silencing of FLC by polycomb proteins, thereby preventing the enrichment of histone 3 lysine 27 trimethylation and eliminating the parental retention of winter cold memory during early embryogenesis [30].
3. The Effect of LEC2 on the Maturation of Plant Seeds
In the maturation stage after embryogenesis, certain storage products are accumulated during the seed filling process in order for growth to be restored under favorable environmental conditions [35][36]. The seed has three different regions: the filial embryo, the filial endosperm, and the maternal seed coat, which have major differences in terms of their genotypes [37]. In various plant species, fatty acids, sugars, starch, and storage proteins accumulate in the endosperm or embryo of the seeds [38]. LEC2 imparts a regulatory effect on the formation of storage compounds during plant seed development. Studies have shown that FUS3 in the L-AFL family could inhibit the expression of GA3ox1 and GA3ox2 (GA biosynthesis genes) [39][40], while LEC2 could activate LEC1 and FUS3 genes to induce embryo maturation [24]. In addition, LEC2 directly induces AGL15 (AGAMOUS-LIKE15) [14], and AGL15 regulates the expression of the GA-related genes GA3ox2 and GA2ox6 [41][42]. These findings indicate that LEC2 regulates genes that are related to GA biosynthesis to affect the embryonic maturation stage of seeds.
In A. thaliana, the main storage compounds of seeds are lipids and seed storage proteins (SSP) [43]. Studies of the regulation of gene expression in plants have demonstrated that SSP is strictly regulated in time and space. Previous research has shown that AtLEC2 regulates the expression of SSP genes. At2S3 is a storage protein gene. Through yeast hybrid screening, the TFs LEC2 and FUS3 were shown to directly activate the expression of the At2S3 promoter and regulate it in a partially redundant manner [11]. Moreover, LEC2 also has a regulatory effect on fatty acid metabolism, mainly because it could influence the WRINKLED1 (WRI1) factor that encodes the transcription of fatty acids [44].
There has been some progress in understanding the mechanism of LEC2 in the regulation of lipids and oil in seeds. Lipids and oils extracted from plants are extremely important renewable bioenergy materials. In plants, the main component of oil in seeds is triacylglycerol (TAG), and its synthesis can be enhanced by artificial modification. Meanwhile, the synthesis of fatty acids can also be artificially regulated. A recent study discovered an interesting phenomenon in A. thaliana: LEC2 could trigger the stockpiling of oil in leaves and seed-specific mRNA. OLEOSIN, the main structural protein of oil bodies during seed development, is highly expressed in seeds [45]. A previous study demonstrated that the effective expression of OLEOSIN in A. thaliana requires the activation of two adjacent RY elements of the LEC2 promoter [46]. LEC2 acts synergistically with ABI3 and LEC1 to enhance the activation of the OLEOSIN promoter in the developing embryo [47]. In summary, LEC2 regulates many genes that participate in different events and signaling pathways of early embryonic development and seed maturation (Figure 1).
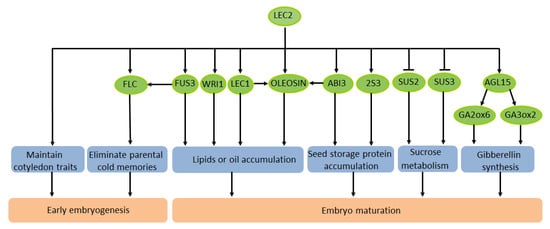
Figure 1. Schematic diagram of the transcription factor LEC2 regulating seed development of A. thaliana.
4. The LEC2 Gene Plays a Crucial Role in Somatic Embryogenesis
Plant cells exhibit unique developmental plasticity that is related to totipotency. For example, the occurrence of somatic embryos is a good indicator of the pluripotency of plant cells. The formation of somatic embryos can be induced by treating cultured somatic cells with auxins and inducing the cells to differentiate in vitro [48]. Numerous studies have shown that the ectopic expression of TFs and their associated genes could induce spontaneous embryogenesis [49][50][51], among which the TF LEC2 is instrumental in inducing the formation of somatic embryos.
In A. thaliana, the LEC2 gene was cloned and expressed ectopically, and the results revealed that it was preferentially expressed during embryogenesis and that it has the ability to induce the formation of somatic embryos [12]. It has been suggested that auxin-induced plant somatic embryogenesis is a key factor in somatic embryo formation, i.e., LEC2 may affect the occurrence of somatic embryos by regulating auxin [52]. A previous study has suggested that AtLEC2 could activate auxin in response to the expression of the INDOLE-3-ACETIC ACID INDUCIBLE30 (IAA30) gene during embryogenesis [14]. Shortly afterward, when LEC2 was ectopically expressed in Arabidopsis seedlings, somatic embryos were formed in the seedlings, and AtLEC2 also activated the expression of the YUCCA2 (YUC2) and YUCCA4 (YUC4) genes for auxin biosynthesis [24]. The above findings imply that the ability of LEC2 to induce the formation of somatic embryos may be derived from the activation of the YUC2 and YUC4 genes that mediate auxin biosynthesis, and that LEC2 acts as a negative regulator of the auxin signal transduction-related IAA30 gene [41][50]. Although the overexpression of LEC2 in plants could induce the formation of somatic embryos, explants treated with auxin in vitro have produced damaged embryos. To better understand this phenomenon, 35S::LEC2-GR transgenic explants were treated with different concentrations of auxin. The results demonstrated that AtLEC2 augments endogenous auxin in the cultured explants, and the expression of three YUCCA genes (YUC1, YUC4, and YUC10) in the IPA-YUC auxin biosynthesis pathway related to somatic embryo induction also showed some correlation with AtLEC2 [53]. Through RT-PCR analysis of the embryogenesis cultures of the explants described above, AtLEC2 was found to be a key regulator that could stimulate the transcription of the YUC1, YUC4, and YUC10 genes. The overexpression of AtLEC2 could also significantly upregulate the expression levels of these three genes when explants were cultured in an auxin-free medium. The increase in endogenous auxin is due to the activation of the YUC gene that regulates the presence and function of exogenous auxin. These findings provide an important perspective for the study of the LEC2-mediated formation of somatic embryos [54].
5. The Function of LEC2 during Other Plant Developmental Stages
LEC2 acts as the master regulatory factor in the processes of plant growth and development. It influences the occurrence of somatic embryos and also plays an important role in the growth phase of other plant structures. Studies have shown that LEC2 is also closely related to the formation of lateral roots. LEC2 activated the transcription of the NAC gene family. NAC proteins play roles in plant developmental processes such as lateral root development [55]. AtLEC2 also interacts with AtFUS3 to activate the expression of the auxin biosynthesis gene YUCCA4 (YUC4), which in turn promotes the generation of lateral roots in A. thaliana [29]. LEC2 and FUS3 have different binding sites in the YUC4 promoter. They can both directly bind to different RY elements of the YUC4 promoter. The FUS3–LEC2 interaction may enhance the ability of binding to RY elements to synergistically activate YUC4 transcription. In the initial stages of lateral root formation, AtLEC2 also activates AtFUS3 expression [29]. The lateral root formation induced by LEC2 was partially attributable to the enhanced FUS3 expression.
In addition, AtLEC2 could induce leaf reprogramming during development. The overexpression of the LEC2 gene in Arabidopsis resulted in alterations of the morphological characteristics of leaves [26]. The leaves became smaller and curled, and developed into cotyledons. Furthermore, the leaves were less fleshy, and the number of trichomes was reduced. Based on the lack of research at the cellular level, this phenomenon could not be further analyzed. After the leaves were sectioned and stained with toluidine blue-O (TBO) dye solution, changes in the anatomical structure of the leaves were assessed. Leaf cells showed a tighter arrangement, and vacuoles were sharply decreased in number and were lightly stained by the dye solution [26].
This entry is adapted from the peer-reviewed paper 10.3390/genes12121896
References
- Niu, D.; He, Y. LEAFY COTYLEDONs: Old genes with new roles beyond seed development. F1000Research 2019, 8, 2144.
- Carbonero, P.; Iglesias-Fernandez, R.; Vicente-Carbajosa, J. The AFL subfamily of B3 transcription factors: Evolution and function in angiosperm seeds. J. Exp. Bot. 2017, 68, 871–880.
- Cagliari, A.; Turchetto-Zolet, A.C.; Korbes, A.P.; Maraschin Fdos, S.; Margis, R.; Margis-Pinheiro, M. New insights on the evolution of leafy cotyledon1 (LEC1) type genes in vascular plants. Genomics 2014, 103, 380–387.
- Lotan, T.; Ohto, M.; Yee, K.M.; West, M.A.; Lo, R.; Kwong, R.W.; Yamagishi, K.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 1998, 93, 1195–1205.
- Kwong, R.W.; Bui, A.Q.; Lee, H.; Kwong, L.W.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 2003, 15, 5–18.
- Hilioti, Z.; Ganopoulos, I.; Bossis, I.; Tsaftaris, A. LEC1-LIKE paralog transcription factor: How to survive extinction and fit in NF-Y protein complex. Gene 2014, 543, 220–233.
- Jia, H.; McCarty, D.R.; Suzuki, M. Distinct roles of LAFL network genes in promoting the embryonic seedling fate in the absence of VAL repression. Plant Physiol. 2013, 163, 1293–1305.
- Vicente-Carbajosa, J.; Carbonero, P. Seed maturation: Developing an intrusive phase to accomplish a quiescent state. Int. J. Dev. Biol. 2005, 49, 645–651.
- Braybrook, S.A.; Harada, J.J. LECs go crazy in embryo development. Trends Plant Sci. 2008, 13, 624–630.
- Harada, J.J. Role of Arabidopsis LEAFY COTYLEDON genes in seed development. J. Plant Physiol. 2001, 158, 405–409.
- Kroj, T.; Savino, G.; Valon, C.; Giraudat, J.; Parcy, F. Regulation of storage protein gene expression in Arabidopsis. Development 2003, 130, 6065–6073.
- Stone, S.L.; Kwong, L.W.; Yee, K.M.; Pelletier, J.; Lepiniec, L.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. USA 2001, 98, 11806–11811.
- Han, J.; Li, X.; Jiang, C.; Wong, G.K.; Rothfels, C.J.; Rao, G. Evolutionary analysis of the LAFL genes involved in the land plant seed maturation program. Front. Plant Sci. 2017, 8, 439.
- Braybrook, S.A.; Stone, S.L.; Park, S.; Bui, A.Q.; Le, B.H.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proc. Natl. Acad. Sci. USA 2006, 103, 3468–3473.
- Jia, H.; Suzuki, M.; McCarty, D.R. Structural variation affecting DNA backbone interactions underlies adaptation of B3 DNA binding domains to constraints imposed by protein architecture. Nucleic Acids Res. 2021, 49, 4989–5002.
- Leviczky, T.; Molnar, E.; Papdi, C.; Oszi, E.; Horvath, G.V.; Vizler, C.; Nagy, V.; Pauk, J.; Bogre, L.; Magyar, Z. E2FA and E2FB transcription factors coordinate cell proliferation with seed maturation. Development 2019, 146, dev179333.
- Tang, X.R.; Bian, S.M.; Tang, M.J.; Lu, Q.; Li, S.B.; Liu, X.G.; Tian, G.; Nguyen, V.; Tsang, E.W.T.; Wang, A.M.; et al. MicroRNA-mediated repression of the seed maturation program during vegetative development in Arabidopsis. PLoS Genet. 2012, 8, e1003091.
- Willmann, M.R.; Mehalick, A.J.; Packer, R.L.; Jenik, P.D. MicroRNAs regulate the timing of embryo maturation in Arabidopsis. Plant Physiol. 2011, 155, 1871–1884.
- Nodine, M.D.; Bartel, D.P. MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis. Genes Dev. 2010, 24, 2678–2692.
- Gutzat, R.; Borghi, L.; Fuetterer, J.; Bischof, S.; Laizet, Y.; Hennig, L.; Feil, R.; Lunn, J.; Gruissem, W. RETINOBLASTOMA-RELATED PROTEIN controls the transition to autotrophic plant development. Development 2011, 138, 2977–2986.
- Meinke, D.W.; Franzmann, L.H.; Nickle, T.C.; Yeung, E.C. Leafy Cotyledon mutants of Arabidopsis. Plant Cell 1994, 6, 1049–1064.
- Gaj, M.D.; Zhang, S.; Harada, J.J.; Lemaux, P.G. Leafy cotyledon genes are essential for induction of somatic embryogenesis of Arabidopsis. Planta 2005, 222, 977–988.
- Mendoza, M.S.; Dubreucq, B.; Miquel, M.; Caboche, M.; Lepiniec, L. LEAFY COTYLEDON 2 activation is sufficient to trigger the accumulation of oil and seed specific mRNAs in Arabidopsis leaves. FEBS Lett. 2005, 579, 4666–4670.
- Stone, S.L.; Braybrook, S.A.; Paula, S.L.; Kwong, L.W.; Meuser, J.; Pelletier, J.; Hsieh, T.F.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity: Implications for somatic embryogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 3151–3156.
- Angeles-Nunez, J.G.; Tiessen, A. Mutation of the transcription factor LEAFY COTYLEDON 2 alters the chemical composition of Arabidopsis seeds, decreasing oil and protein content, while maintaining high levels of starch and sucrose in mature seeds. J. Plant Physiol. 2011, 168, 1891–1900.
- Feeney, M.; Frigerio, L.; Cui, Y.; Menassa, R. Following vegetative to embryonic cellular changes in leaves of Arabidopsis overexpressing LEAFY COTYLEDON2. Plant Physiol. 2013, 162, 1881–1896.
- Kim, H.U.; Jung, S.J.; Lee, K.R.; Kim, E.H.; Lee, S.M.; Roh, K.H.; Kim, J.B. Ectopic overexpression of castor bean LEAFY COTYLEDON2 (LEC2) in Arabidopsis triggers the expression of genes that encode regulators of seed maturation and oil body proteins in vegetative tissues. FEBS Open Bio 2013, 4, 25–32.
- Iwase, A.; Mita, K.; Nonaka, S.; Ikeuchi, M.; Koizuka, C.; Ohnuma, M.; Ezura, H.; Imamura, J.; Sugimoto, K. WIND1-based acquisition of regeneration competency in Arabidopsis and rapeseed. J. Plant Res. 2015, 128, 389–397.
- Tang, L.P.; Zhou, C.; Wang, S.S.; Yuan, J.; Zhang, X.S.; Su, Y.H. FUSCA 3 interacting with LEAFY COTYLEDON 2 controls lateral root formation through regulating YUCCA4 gene expression in Arabidopsis thaliana. New Phytol. 2017, 213, 1740–1754.
- Tao, Z.; Hu, H.; Luo, X.; Jia, B.; Du, J.; He, Y. Embryonic resetting of the parental vernalized state by two B3 domain transcription factors in Arabidopsis. Nat. Plants 2019, 5, 424–435.
- Barreto, H.G.; Ságio, S.A.; Chalfun-Júnior, A.; Fevereiro, P.; Benedito, V.A. Transcriptional profiling of the AFL subfamily of B3-type transcription factors during the in vitro induction of somatic embryogenesis in the model legume Medicago truncatula. Plant Cell Tissue Organ Cult. 2019, 139, 327–337.
- Meinke, D.W. A homoeotic mutant of Arabidopsis thaliana with leafy cotyledons. Science 1992, 258, 1647–1650.
- Qüesta, J.I.; Song, J.; Geraldo, N.; An, H.; Dean, C. Arabidopsis transcriptional repressor VAL1 triggers Polycomb silencing at FLC during vernalization. Science 2016, 353, 485–488.
- Yuan, W.; Luo, X.; Li, Z.; Yang, W.; Wang, Y.; Liu, R.; Du, J.; He, Y. A cis cold memory element and a trans epigenome reader mediate Polycomb silencing of FLC by vernalization in Arabidopsis. Nat. Genet. 2016, 48, 1527–1534.
- McCarty, D.R. Genetic control and integration of maturation and germination pathways in seed development. Annu. Rev. Plant Biol. 1995, 46, 71–93.
- Goldberg, R.B.; Paiva, G.; Yadegari, R. Plant embryogenesis: Zygote to seed. Science 1994, 266, 605–614.
- Jo, L.; Pelletier, J.M.; Harada, J.J. Central role of the LEAFY COTYLEDON1 transcription factor in seed development. J. Integr. Plant. Biol. 2019, 61, 564–580.
- Higgins, T. Synthesis and regulation of major proteins in seeds. Annu. Rev. Plant Phys. 1984, 35, 191–221.
- Curaba, J.; Moritz, T.; Blervaque, R.; Parcy, F.; Raz, V.; Herzog, M.; Vachon, G. AtGA3ox2, a key gene responsible for bioactive gibberellin biosynthesis, is regulated during embryogenesis by LEAFY COTYLEDON2 and FUSCA3 in Arabidopsis. Plant Physiol. 2004, 136, 3660–3669.
- Gazzarrini, S.; Tsuchiya, Y.; Lumba, S.; Okamoto, M.; McCourt, P. The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev. Cell 2004, 7, 373–385.
- Kumar, V.; van Staden, J. New insights into plant somatic embryogenesis: An epigenetic view. Acta Physiol. Plant 2017, 39, 194.
- Wang, H.; Caruso, L.V.; Downie, A.B.; Perry, S.E. The embryo MADS domain protein AGAMOUS-Like 15 directly regulates expression of a gene encoding an enzyme involved in gibberellin metabolism. Plant Cell 2004, 16, 1206–1219.
- Fait, A.; Angelovici, R.; Less, H.; Ohad, I.; Urbanczyk-Wochniak, E.; Fernie, A.R.; Galili, G. Arabidopsis seed development and germination is associated with temporally distinct metabolic switches. Plant Physiol. 2006, 142, 839–854.
- Baud, S.; Mendoza, M.S.; To, A.; Harscoet, E.; Lepiniec, L.; Dubreucq, B. WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J. 2007, 50, 825–838.
- Huang, A.H. Oleosins and oil bodies in seeds and other organs. Plant Physiol. 1996, 110, 1055–1061.
- Che, N.; Yang, Y.; Li, Y.; Wang, L.; Huang, P.; Gao, Y.; An, C. Efficient LEC2 activation of OLEOSIN expression requires two neighboring RY elements on its promoter. Sci. China C Life Sci. 2009, 52, 854–863.
- Baud, S.; Kelemen, Z.; Thevenin, J.; Boulard, C.; Blanchet, S.; To, A.; Payre, M.; Berger, N.; Effroy-Cuzzi, D.; Franco-Zorrilla, J.M.; et al. Deciphering the molecular mechanisms underpinning the transcriptional control of gene expression by master transcriptional regulators in Arabidopsis seed. Plant Physiol. 2016, 171, 1099–1112.
- Feher, A. Somatic embryogenesis—Stress-induced remodeling of plant cell fate. BBA-Gene Regul. Mech. 2015, 1849, 385–402.
- Horstman, A.; Bemer, M.; Boutilier, K. A transcriptional view on somatic embryogenesis. Regeneration 2017, 4, 201–216.
- Jha, P.; Kumar, V. BABY BOOM (BBM): A candidate transcription factor gene in plant biotechnology. Biotechnol. Lett. 2018, 40, 1467–1475.
- Salvo, S.A.; Hirsch, C.N.; Buell, C.R.; Kaeppler, S.M.; Kaeppler, H.F. Whole transcriptome profiling of maize during early somatic embryogenesis reveals altered expression of stress factors and embryogenesis-related genes. PLoS ONE 2014, 9, e111407.
- Gaj, M.D. Factors influencing somatic embryogenesis induction and plant regeneration with particular reference to Arabidopsis thaliana (L.) Heynh. Plant Growth Regul. 2004, 43, 27–47.
- Wojcikowska, B.; Jaskola, K.; Gasiorek, P.; Meus, M.; Nowak, K.; Gaj, M.D. LEAFY COTYLEDON2 (LEC2) promotes embryogenic induction in somatic tissues of Arabidopsis, via YUCCA-mediated auxin biosynthesis. Planta 2013, 238, 425–440.
- Kumar, V.; Jha, P.; van Staden, J. LEAFY COTYLEDONs (LECs): Master regulators in plant embryo development. Plant Cell Tissue Organ Cult. 2020, 140, 475–487.
- He, X.J.; Mu, R.L.; Cao, W.H.; Zhang, Z.G.; Zhang, J.S.; Chen, S.Y. AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 2005, 44, 903–916.
This entry is offline, you can click here to edit this entry!
