Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Cell Biology
PΦB is an open-chain tetrapyrrole chromophore, a critical synthetase for phytochromes to function as a light receptor to regulate plant growth and development. Arabidopsis HY2 encodes a key synthase of PΦB, which is a ferredoxin-dependent biliverdin reductase that catalyzes the reduction in the A-ring 2,3,31,32-diene system to produce an ethylidene group for assembly with apophytochromes.
- Arabidopsis
- HY2
- salt stress
- seed germination
1. Introduction
Saline soil is an unfavorable environmental factor that seriously affects seed germination, seedling growth, and even final yield in crops [1][2][3]. About 7% of the total land surface and 20% of the irrigated land are affected by soil with excessive salt concentration, and the situation is getting worse [4][5]. Global climate change and poor irrigation water quality are the main factors leading to soil salinization [6][7]. Therefore, it is urgent to study the molecular mechanism of plants’ adaption to saline soil and to enhance such adaptability of plants to saline soil through molecular genetic improvement.
As salt stress gives rise to ion stress, osmotic stress, secondary stress, and oxidative stress [8][9], it is crucial for plants to maintain the balance between ion, osmosis, and ROS. In addition, plants have evolved a series of mechanisms to maintain salt balance in the long process of evolution [10]. In terms of ionic stress, after the perception of a salt-stress signal induced by a high concentration of salt in plants, the salt receptor glycosyl inositol phosphoryl ceramide (GIPC) [11][12] can directly bind to the external Na+ to form a direct interaction, which activates the Ca2+ channel. The influx of Ca2+ thus drives the adaptive response to high salt levels, which promotes EF-hand Ca2+-binding proteins SOS3 to activate serine/threonine protein kinase SOS2, and then to activate Na+/H+ antiporter SOS1 to pump Na+ out of the cell, maintaining the salinity balance in vivo [13][14]. In terms of osmotic stress, the synthesis of compatible osmolytes is crucial for the maintenance of osmotic potential and protein structure in cells, including the expression of related genes as PM-located protein OSCA, MAPK cascades, SnRK2 isoforms, etc. [15][16][17], and the synthesis of the accumulation of related substances, such as proline, betaine, sugars, etc. [18][19][20]. In terms of oxidative stress, both the gene expressions, such as MAPKKK–MAPKK–MAPK (mitogen-activated protein kinase) cascade [21], and the ROS scavenging enzymes, such as superoxide dismutase (SOD), peroxidase (POD), catalase (CAT), etc. [22], act to maintain the balance of ROS in vivo. Therefore, improving the molecular mechanism of salt stress is very important for the strengthening of salt tolerance in plants.
PΦB is an open-chain tetrapyrrole chromophore, a critical synthetase for phytochromes to function as a light receptor to regulate plant growth and development [23][24]. Arabidopsis HY2 encodes a key synthase of PΦB [25][26], which is a ferredoxin-dependent biliverdin reductase that catalyzes the reduction in the A-ring 2,3,31,32-diene system to produce an ethylidene group for assembly with apophytochromes [27]. Furthermore, it has been reported that HY2 induces the synthesis of phyA to inhibit the elongation of hypocotyl under the far-red light treatment [28]. Under the treatment of exogenous trehalose, the expression of HY2 is upregulated by 2.8 times [29]. Besides, HY2 participates in the apoplastic and chloroplastic redox signaling networks, being responsible for chlorophyll biosynthesis [30]. However, whether HY2 is involved in the plant stress response signal network remains unknown.
2. Disruption of HY2 Reduces, and Overexpression of HY2 Enhances, NaCl Sensitivity during Seed Germination
To analyze the novel salt tolerance genes, we used a luciferase reporter system to construct different Arabidopsis transgene lines overexpressing firefly luciferase (LUC). Compared with col4 and LUC-vector lines, the lines expressing the PΦB synthetase HY2 showed a salt-hypersensitive phenotype and hy2 mutant displayed a salt-resistant phenotype with 200 mM NaCl stress (Figure 1A). We found most hy2 mutant seedlings germinated 3 days after being sown in the medium containing 200 mM NaCl, while the seedlings of col4 and LUC-vector needed 4–5 days to germinate; compared with col4, the germination rate of lines overexpressing LUC–HY2 was significantly lower, but that of hy2 mutant was obviously higher (Figure 1B).
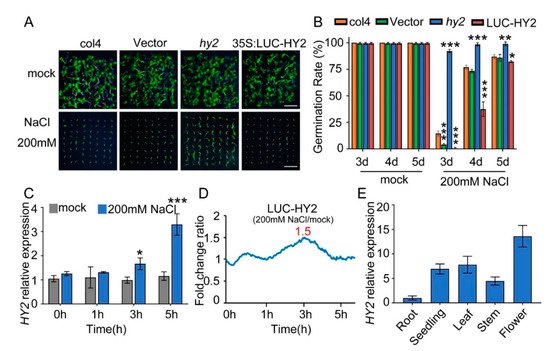
Figure 1. NaCl sensitivity of hy2 mutant and HY2-overexpressing lines. (A) Phenotypic comparison. Col4, LUC-vector, hy2 mutant, and LUC–HY2 overexpression lines were sown, respectively, on 1/2 MS medium (as mock) or 1/2 MS medium containing indicated concentrations of 200 mM NaCl for 4 d. Scare bar: 1 cm. (B) Seed germination assay. Seeds were transferred to 1/2 MS containing 200 mM NaCl, and then the seed germination rate was calculated at 3–5 d. Data are shown as mean ± SD (n = 3). More than 64 seeds were measured in each replicate. (C) QPCR analysis of HY2 expression in 5-day-old col4 seedlings treated with or without 200 mM NaCl for 0–5 h. Data are shown as mean ± SD (n = 3). (D) LUC signals in 5-day-old LUC–HY2 overexpression line seedlings treated with or without 200 mM NaCl. Signals were detected every 10 min, and the detecting period was 5 h. (E) QPCR analysis of HY2 expression in different tissues of Arabidopsis. Data are shown as mean ± SD (n = 3). Asterisks in (B,C) indicate statistically significant differences compared with relevant col4 plants: * p < 0.05, ** p < 0.01, *** p < 0.001.
The following QPCR analysis showed that, with 0 h and 1 h NaCl stress, the transcription level of HY2 was undifferentiated; with 3 h and 5 h NaCl stress, the transcription level of HY2 increased by 1.7 and 2.8 times, respectively (Figure 1C). The luciferase assay showed a 1.5-fold increase in the protein level of HY2 after 3 h NaCl stress (Figure 1D). These results indicated that NaCl significantly mediates and upregulates the HY2, both on its transcription level and protein level. At the same time, we analyzed the expression of HY2 in different tissues, showing that HY2 was expressed in different tissues of Arabidopsis, but its expression level was the lowest in roots and the highest in flowers (Figure 1E).
3. Disruption of HY2-Altered Expression of a Set of Stress-Responsive Genes
NaCl stress is one of the stress factors that affects the growth and development of seeds, and induces the expression of stress-related proteins, such as seed growth and development related proteins and ABA-pathway-related proteins. Therefore, we verified the expression patterns of proteins related to salt stress, seed growth and development, and ABA pathway in col4 and hy2 mutant (Figure 7A), from which we found that 15 proteins were mediated by NaCl stress but not by HY2 in a specific way (gray dot), while 32 were specifically mediated by HY2 (green and yellow dot). All of the proteins were then divided into different groups: proteins involved in the salt stress, proteins participating in the ABA pathway, and proteins taking part in the growth and development of seeds, among which the number of proteins specifically regulated by HY2 (col4-specific and hy2-specific) were 22, 10, and 5, respectively, while the number of proteins specifically regulated by NaCl stress were only 9, 7, and 1 (less than half the number of proteins specifically regulated by HY2), respectively. These results showed, under NaCl stress conditions, that proteins involved in the salt stress pathway dominated, accounting for 66%, and those involved in the ABA pathway and seed growth/development took the secondary place (Figure 7B), indicating that HY2 simultaneously regulates the protein expression related to salt stress, ABA pathway, and seed growth and development. As HY2 is a potential regulator involved in NaCl signaling (Figure 1A), the expression of stress inductive genes, such as RD29A, RD29B, and DREB2A, was tested in hy2 mutant. We tested the inducible genes under the conditions of 0 h–5 h stress of 200 mM NaCl, and found that the expression levels of RD29A and RD29B were undifferentiated at 0 h and 1 h in hy2 mutant, but significantly decreased at 3 h and 5 h when compared with those in col4 at the same conditions (Figure 7C,D); the expression level of DREB2A remained the same at 0 h, 1 h, and 5 h, and was significantly downregulated at 3 h only (Figure 7E), indicating that HY2 induces the expression of NaCl inducible genes and positively regulates NaCl signaling.
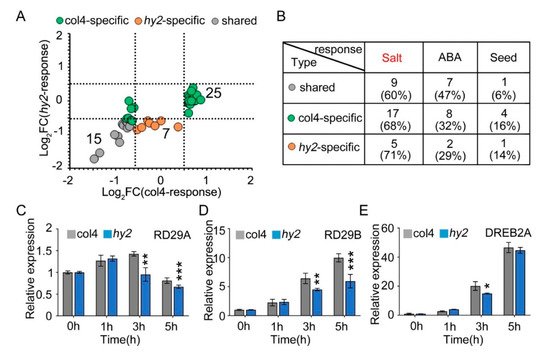
Figure 7. The expression of stress-responsive genes. (A) Protein expression pattern of col4 and hy2 shared proteins involved in salt, ABA, and seed-related pathway. Color denotes proteins regulated similarly by col4 and hy2 (grey), or specifically by col4 (green) or hy2 (orange). (B) The number and proportion of col4 and hy2 shared proteins involved in salt, ABA, and seed-related pathway. (C–E) The expression of RD29A, RD29B, and DREB2A in col4 and hy2 mutant seedlings treated with exogenous NaCl. The 5-day-old col4 and hy2 mutant seedlings were transferred to 1/2 MS solution with or without 200 mM NaCl for 0–5 h, and then the seedlings were harvested for QPCR. Data are shown as mean ± SD (n = 3). Asterisks showed in (B–D) indicate statistically significant differences compared with relevant col4 plants: * p < 0.05, ** p < 0.01, *** p < 0.001.
4. Interaction Network of HY2-Specific DRPs
Protein interaction networks were generated to evaluate the interaction of the DRPs (known and unknown proteins) specifically regulated by HY2 (Figure 8). We selected 22 proteins specifically regulated by HY2 under salt stress, among which 17 were col4-specific (14 upregulated and 3 downregulated) and five were hy2-specific (five downregulated). The results showed that ANNAT1–ANNAT2–ANNAT3–ANNAT4, the family members of Annexins (a family of calcium dependent membrane binding proteins), were involved in salt-stress responses specifically regulated by HY2 in a mutual-functioning way. The score of association between ANNAT1 and ANNAT2 was 0.915, which was the highest, while that between ANNAT1 and ANNAT4 was 0.805, which was the lowest. Glutathione transferase GSTU19 induced by drought, oxidative stress, and hormonal responses; Glutathione S-transferase PHI 10 (GSTF10) involved in water deprivation and toxin catabolic process; 60S ribosomal protein L5 (RPL5A, RPL5B) responsible for the synthesis of proteins in the cell as the component of the ribosome; and ribosomal proteins of L7Ae/L30e/S12e/Gadd45 family protein (AT2G32060) involved in translation all interactionally participated in salt-stress responses specifically regulated by HY2. The association score between AT2G32060 and RPL5A/RPL5B was 0.999, while that between GSTF10 and RPL5A/RPL5B was 0.596. We, therefore, identified two interaction protein networks specifically regulated by HY2 under salt stress.
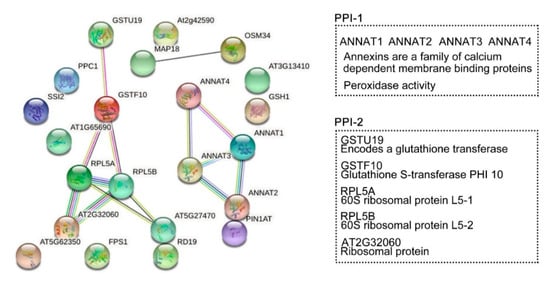
Figure 8. Protein-protein interaction (PPI) networks. STRING analysis using the HY2-specific regulated proteins under salt stress. Network nodes represent proteins. Edges represent protein–protein associations. Known interactions:  from curated databases;
from curated databases;  experimentally determined. Predicted interactions:
experimentally determined. Predicted interactions:  protein neighborhood;
protein neighborhood;  protein fusions;
protein fusions;  protein co-occurrence. Others:
protein co-occurrence. Others:  textmining;
textmining;  co-expression;
co-expression;  protein homology. Tow PPI specifically regulated by HY2. PPI-1: ANNAT1–ANNAT2–ANNAT3–ANNAT4. PPI-2: GSTU19–GSTF10–RPL5A–RPL5B–AT2G32060.
protein homology. Tow PPI specifically regulated by HY2. PPI-1: ANNAT1–ANNAT2–ANNAT3–ANNAT4. PPI-2: GSTU19–GSTF10–RPL5A–RPL5B–AT2G32060.
 from curated databases;
from curated databases;  experimentally determined. Predicted interactions:
experimentally determined. Predicted interactions:  protein neighborhood;
protein neighborhood;  protein fusions;
protein fusions;  protein co-occurrence. Others:
protein co-occurrence. Others:  textmining;
textmining;  co-expression;
co-expression;  protein homology. Tow PPI specifically regulated by HY2. PPI-1: ANNAT1–ANNAT2–ANNAT3–ANNAT4. PPI-2: GSTU19–GSTF10–RPL5A–RPL5B–AT2G32060.
protein homology. Tow PPI specifically regulated by HY2. PPI-1: ANNAT1–ANNAT2–ANNAT3–ANNAT4. PPI-2: GSTU19–GSTF10–RPL5A–RPL5B–AT2G32060.References
- Morton, M.J.L.; Awlia, M.; Al-Tamimi, N.; Saade, S.; Pailles, Y.; Negrão, S.; Tester, M. Salt stress under the scalpel—Dissecting the genetics of salt tolerance. Plant J. Cell Mol. Biol. 2019, 97, 148–163.
- Acosta-Motos, J.R.; Ortuño, M.F.; Bernal-Vicente, A.; Diaz-Vivancos, P.; Sanchez-Blanco, M.J.; Hernandez, J.A. Plant responses to salt stress: Adaptive mechanisms. Agronomy 2017, 7, 18.
- Shahid, M.A.; Sarkhosh, A.; Khan, N.; Balal, R.M.; Ali, S.; Rossi, L.; Gómez, C.; Mattson, N.; Nasim, W.; Garcia-Sanchez, F.J.A. Insights into the physiological and biochemical impacts of salt stress on plant growth and development. Agronomy 2020, 10, 938.
- Yuan, F.; Leng, B.; Wang, B. Progress in Studying Salt Secretion from the Salt Glands in Recretohalophytes: How Do Plants Secrete Salt? Front. Plant Sci. 2016, 7, 977.
- Yang, Z.; Li, J.L.; Liu, L.N.; Xie, Q.; Sui, N. Photosynthetic Regulation Under Salt Stress and Salt-Tolerance Mechanism of Sweet Sorghum. Front. Plant Sci. 2019, 10, 1722.
- Vaishnav, A.; Shukla, A.K.; Sharma, A.; Kumar, R.; Choudhary, D.K. Endophytic bacteria in plant salt stress tolerance: Current and future prospects. J. Plant Growth Regul. 2019, 38, 650–668.
- Al-Taey, D.K.; Al-Musawi, Z.J. Effect of Nano-fertilizers, salicylic acid, and organic matter in growth and yield of rocket (Eruca sativa Mill) under Salt stress. Int. J. Bot. Stud. 2019, 4, 77–81.
- Yang, Y.; Guo, Y. Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 2018, 60, 796–804.
- Yang, Y.; Guo, Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2018, 217, 523–539.
- Sun, Y.; Zhao, J.; Li, X.; Li, Y. E2 conjugases UBC1 and UBC2 regulate MYB42-mediated SOS pathway in response to salt stress in Arabidopsis. New Phytol. 2020, 227, 455–472.
- Jiang, Z.; Zhou, X.; Tao, M.; Yuan, F.; Liu, L.; Wu, F.; Wu, X.; Xiang, Y.; Niu, Y.; Liu, F.; et al. Plant cell-surface GIPC sphingolipids sense salt to trigger Ca(2+) influx. Nature 2019, 572, 341–346.
- Steinhorst, L.; Kudla, J. How plants perceive salt. Nature 2019, 572, 318–320.
- Ma, L.; Ye, J.; Yang, Y.; Lin, H.; Yue, L.; Luo, J.; Long, Y.; Fu, H.; Liu, X.; Zhang, Y.; et al. The SOS2-SCaBP8 Complex Generates and Fine-Tunes an AtANN4-Dependent Calcium Signature under Salt Stress. Dev. Cell 2019, 48, 697–709.e695.
- Chai, H.; Guo, J.; Zhong, Y.; Hsu, C.C.; Zou, C.; Wang, P.; Zhu, J.K.; Shi, H. The plasma-membrane polyamine transporter PUT3 is regulated by the Na(+)/H(+) antiporter SOS1 and protein kinase SOS2. New Phytol. 2020, 226, 785–797.
- Yuan, F.; Yang, H.; Xue, Y.; Kong, D.; Ye, R.; Li, C.; Zhang, J.; Theprungsirikul, L.; Shrift, T.; Krichilsky, B.; et al. OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature 2014, 514, 367–371.
- Gasulla, F.; Barreno, E.; Parages, M.L.; Cámara, J.; Jiménez, C.; Dörmann, P.; Bartels, D. The Role of Phospholipase D and MAPK Signaling Cascades in the Adaption of Lichen Microalgae to Desiccation: Changes in Membrane Lipids and Phosphoproteome. Plant Cell Physiol. 2016, 57, 1908–1920.
- Zhu, J.K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324.
- Naliwajski, M.; Skłodowska, M. The Relationship between the Antioxidant System and Proline Metabolism in the Leaves of Cucumber Plants Acclimated to Salt Stress. Cells 2021, 10, 609.
- Yamada, N.; Takahashi, H.; Kitou, K.; Sahashi, K.; Tamagake, H.; Tanaka, Y.; Takabe, T. Suppressed expression of choline monooxygenase in sugar beet on the accumulation of glycine betaine. Plant Physiol. Biochem. PPB 2015, 96, 217–221.
- Boriboonkaset, T.; Theerawitaya, C.; Yamada, N.; Pichakum, A.; Supaibulwatana, K.; Cha-Um, S.; Takabe, T.; Kirdmanee, C. Regulation of some carbohydrate metabolism-related genes, starch and soluble sugar contents, photosynthetic activities and yield attributes of two contrasting rice genotypes subjected to salt stress. Protoplasma 2013, 250, 1157–1167.
- Pitzschke, A.; Djamei, A.; Bitton, F.; Hirt, H. A major role of the MEKK1-MKK1/2-MPK4 pathway in ROS signalling. Mol. Plant 2009, 2, 120–137.
- Du, C.; Zhao, P.; Zhang, H.; Li, N.; Zheng, L.; Wang, Y. The Reaumuria trigyna transcription factor RtWRKY1 confers tolerance to salt stress in transgenic Arabidopsis. J. Plant Physiol. 2017, 215, 48–58.
- Bianchetti, R.E.; Cruz, A.B.; Oliveira, B.S.; Demarco, D.; Purgatto, E.; Peres, L.E.P.; Rossi, M.; Freschi, L. Phytochromobilin deficiency impairs sugar metabolism through the regulation of cytokinin and auxin signaling in tomato fruits. Sci. Rep. 2017, 7, 7822.
- Hasegawa, J.-Y.; Isshiki, M.; Fujimoto, K.; Nakatsuji, H.J.C.P.L. Structure of phytochromobilin in the Pr and Pfr forms: SAC-CI theoretical study. Chem. Phys. Lett. 2005, 410, 90–93.
- Rockwell, N.C.; Martin, S.S.; Li, F.W.; Mathews, S.; Lagarias, J.C. The phycocyanobilin chromophore of streptophyte algal phytochromes is synthesized by HY2. New Phytol. 2017, 214, 1145–1157.
- Kohchi, T.; Mukougawa, K.; Frankenberg, N.; Masuda, M.; Yokota, A.; Lagarias, J.C. The Arabidopsis HY2 gene encodes phytochromobilin synthase, a ferredoxin-dependent biliverdin reductase. Plant Cell 2001, 13, 425–436.
- Busch, A.W.; Reijerse, E.J.; Lubitz, W.; Frankenberg-Dinkel, N.; Hofmann, E. Structural and mechanistic insight into the ferredoxin-mediated two-electron reduction of bilins. Biochem. J. 2011, 439, 257–264.
- Parks, B.M.; Quail, P.H. Phytochrome-Deficient hy1 and hy2 Long Hypocotyl Mutants of Arabidopsis Are Defective in Phytochrome Chromophore Biosynthesis. Plant Cell 1991, 3, 1177–1186.
- Bae, H.; Herman, E.; Bailey, B.; Bae, H.J.; Sicher, R.J.P.P. Exogenous trehalose alters Arabidopsis transcripts involved in cell wall modification, abiotic stress, nitrogen metabolism, and plant defense. Physiol. Plant. 2005, 125, 114–126.
- Sierla, M.; Rahikainen, M.; Salojärvi, J.; Kangasjärvi, J.; Kangasjärvi, S. Apoplastic and chloroplastic redox signaling networks in plant stress responses. Antioxid. Redox Signal. 2013, 18, 2220–2239.
This entry is offline, you can click here to edit this entry!
