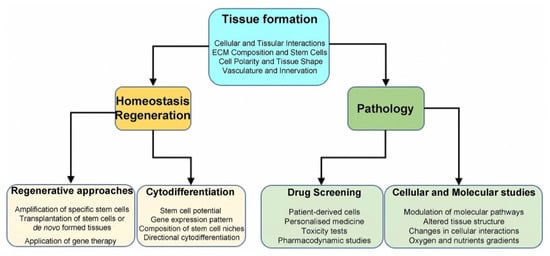
Three-dimensional (3D) culture systems opened up new horizons in the emerging field of regenerative medicine and in studying the biology of tissues and organs, modelling various diseases, and screening drugs. Producing accurate in vitro models increases the possibilities for studying molecular control of cell–cell and cell–microenvironment interactions in detail. The Notch signalling is linked to cell fate determination, tissue definition, and maintenance in both physiological and pathological conditions. Hence, 3D cultures provide new accessible platforms for studying activation and modulation of the Notch pathway.
- spheroids
- organoids
- organ-on-a-chip
- microfluidics
- 3D culture systems
- Notch signalling
- cancer
- drug screening
- regeneration
1. Introduction
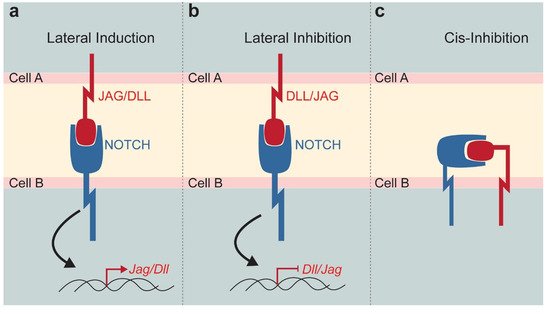
2. The Notch Signalling Pathway
3. 3D Culture Systems
3.1. Spheroids
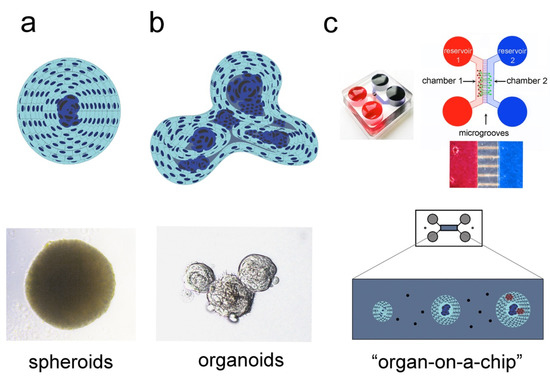
3.2. Organoids
3.3. “Organ-on-a-chip” Technology
4. 3D Systems in Regenerative Medicine
5. 3D Cultures in Disease Modelling
6. Drug Screening and Therapy Testing in 3D Culture Systems
7. Conclusions
Abbreviations
| NICD | Notch Intracellular Domain |
| Dll4 | Delta-like4 |
| 3D | Three-dimensional |
| EBs | Embryoid Bodies |
| PSCs | Pluripotent Stem Cells |
| MSCs | Mesenchymal Stem Cells |
| HIF1 | Hypoxia inducible factor 1 |
| ASCs | Adult Stem Cells |
| ECM | Extracellular Matrix |
| ESCs | Embryonic Stem Cells |
| iPSC | Induced-Pluripotent Stem Cells |
| TNF | Tumour Necrosis Factor |
| IL1 | Interleukin-1 |
| NGS | Next Generation Sequencing |
| DSCs | Dental Stem Cells |
This entry is adapted from the peer-reviewed paper 10.3390/ijms222212473
References
- Jensen, C.; Teng, Y. Is It Time to Start Transitioning From 2D to 3D Cell Culture? Front. Mol. Biosci. 2020, 7, 33.
- Chaicharoenaudomrung, N.; Kunhorm, P.; Noisa, P. Three-dimensional cell culture systems as an in vitro platform for cancer and stem cell modeling. World J. Stem Cells 2019, 11, 1065–1083.
- Doctor, A.; Seifert, V.; Ullrich, M.; Hauser, S.; Pietzsch, J. Three-Dimensional Cell Culture Systems in Radiopharmaceutical Cancer Research. Cancers 2020, 12, 2765.
- Fang, Y.; Eglen, R.M. Three-Dimensional Cell Cultures in Drug Discovery and Development. SLAS Discov. Adv. Life Sci. R D 2017, 22, 456–472.
- Koch, U.; Lehal, R.; Radtke, F. Stem cells living with a Notch. Development 2013, 140, 689–704.
- Mitsiadis, T.A.; Graf, D.; Luder, H.; Gridley, T.; Bluteau, G. BMPs and FGFs target Notch signalling via jagged 2 to regulate tooth morphogenesis and cytodifferentiation. Development 2010, 137, 3025–3035.
- Mitsiadis, T.A.; Henrique, D.; Thesleff, I.; Lendahl, U. Mouse Serrate-1 (Jagged-1): Expression in the developing tooth is regulated by epithelial-mesenchymal interactions and fibroblast growth factor-4. Development 1997, 124, 1473–1483.
- Mitsiadis, T.A.; Lardelli, M.; Lendahl, U.; Thesleff, I. Expression of Notch 1, 2 and 3 is regulated by epithelial-mesenchymal interactions and retinoic acid in the developing mouse tooth and associated with determination of ameloblast cell fate. J. Cell Biol. 1995, 130, 407–418.
- Simpson, P. Lateral inhibition and the development of the sensory bristles of the adult peripheral nervous system of Drosophila. Dev. Camb. Engl. 1990, 109, 509–519.
- Louvi, A.; Artavanis-Tsakonas, S. Notch signalling in vertebrate neural development. Nat. Rev. Neurosci. 2006, 7, 93–102.
- Artavanis-Tsakonas, S. Notch Signaling: Cell Fate Control and Signal Integration in Development. Science 1999, 284, 770–776.
- Lewis, J. Notch signalling and the control of cell fate choices in vertebrates. Semin. Cell Dev. Biol. 1998, 9, 583–589.
- Braune, E.-B.; Lendahl, U. Notch—A goldilocks signaling pathway in disease and cancer therapy. Discov. Med. 2016, 21, 189–196.
- Artavanis-Tsakonas, S.; Delidakis, C.; Fehon, R.G. The Notch locus and the cell biology of neuroblast segregation. Annu. Rev. Cell Biol. 1991, 7, 427–452.
- Bigas, A.; Porcheri, C. Notch and Stem Cells. Adv. Exp. Med. Biol. 2018, 1066, 235–263.
- Irvine, K.D.; Vogt, T.F. Dorsal-ventral signaling in limb development. Curr. Opin. Cell Biol. 1997, 9, 867–876.
- Monterubbianesi, R.; Bencun, M.; Pagella, P.; Woloszyk, A.; Orsini, G.; Mitsiadis, T.A. A comparative in vitro study of the osteogenic and adipogenic potential of human dental pulp stem cells, gingival fibroblasts and foreskin fibroblasts. Sci. Rep. 2019, 9, 1761.
- Mitsiadis, T.A.; Barrandon, O.; Rochat, A.; Barrandon, Y.; De Bari, C. Stem cell niches in mammals. Exp. Cell Res. 2007, 313, 3377–3385.
- Pagella, P.; Vargas Roditi, L.; Stadlinger, B.; Moor, A.E.; Mitsiadis, T.A. Notch signaling in the dynamics of perivascular stem cells and their niches. Stem Cells Transl. Med. 2021, 10, 1433–1445.
- Porcheri, C.; Golan, O.; Calero-Nieto, F.J.; Thambyrajah, R.; Ruiz-Herguido, C.; Wang, X.; Catto, F.; Guillén, Y.; Sinha, R.; González, J.; et al. Notch ligand Dll4 impairs cell recruitment to aortic clusters and limits blood stem cell generation. EMBO J. 2020, 39, e104270.
- Pagella, P.; Miran, S.; Neto, E.; Martin, I.; Lamghari, M.; Mitsiadis, T.A. Human dental pulp stem cells exhibit enhanced properties in comparison to human bone marrow stem cells on neurites outgrowth. FASEB J. 2020, 34, 5499–5511.
- Knoblich, J.A.; Jan, L.Y.; Jan, Y.N. Asymmetric segregation of Numb and Prospero during cell division. Nature 1995, 377, 624–627.
- Gaiano, N.; Nye, J.S.; Fishell, G. Radial glial identity is promoted by Notch1 signaling in the murine forebrain. Neuron 2000, 26, 395–404.
- Knoblich, J.A. Asymmetric cell division during animal development. Nat. Rev. Mol. Cell Biol. 2001, 2, 11–20.
- Ables, J.L.; DeCarolis, N.A.; Johnson, M.A.; Rivera, P.D.; Gao, Z.; Cooper, D.C.; Radtke, F.; Hsieh, J.; Eisch, A.J. Notch1 Is Required for Maintenance of the Reservoir of Adult Hippocampal Stem Cells. J. Neurosci. 2010, 30, 10484–10492.
- Louvi, A.; Artavanis-Tsakonas, S. Notch and disease: A growing field. Semin. Cell Dev. Biol. 2012, 23, 473–480.
- Ho, D.M.; Artavanis-Tsakonas, S. The Notch-Mediated Proliferation Circuitry. Curr. Top. Dev. Biol. 2016, 116, 17–33.
- Reichrath, J.; Reichrath, S. The Impact of Notch Signaling for Carcinogenesis and Progression of Nonmelanoma Skin Cancer: Lessons Learned from Cancer Stem Cells, Tumor Angiogenesis, and Beyond. Adv. Exp. Med. Biol. 2021, 1287, 123–154.
- Stylianou, S.; Clarke, R.B.; Brennan, K. Aberrant activation of notch signaling in human breast cancer. Cancer Res. 2006, 66, 1517–1525.
- Deng, G.; Ma, L.; Meng, Q.; Ju, X.; Jiang, K.; Jiang, P.; Yu, Z. Notch signaling in the prostate: Critical roles during development and in the hallmarks of prostate cancer biology. J. Cancer Res. Clin. Oncol. 2016, 142, 531–547.
- Wilson, H.V. On some phenomena of coalescence and regeneration in sponges. J. Exp. Zool. 1907, 5, 245–258.
- Wilson, H.V. A new method by which sponges may be artificially reared. Science 1907, 25, 912–915.
- Martin, G.R. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. USA 1981, 78, 7634–7638.
- Desai, N.; Rambhia, P.; Gishto, A. Human embryonic stem cell cultivation: Historical perspective and evolution of xeno-free culture systems. Reprod. Biol. Endocrinol. 2015, 13, 9.
- Morgani, S.M.; Canham, M.A.; Nichols, J.; Sharov, A.A.; Migueles, R.P.; Ko, M.S.H.; Brickman, J.M. Totipotent embryonic stem cells arise in ground-state culture conditions. Cell Rep. 2013, 3, 1945–1957.
- Achilli, T.-M.; Meyer, J.; Morgan, J.R. Advances in the formation, use and understanding of multi-cellular spheroids. Expert Opin. Biol. Ther. 2012, 12, 1347–1360.
- Clevers, H. Modeling Development and Disease with Organoids. Cell 2016, 165, 1586–1597.
- Natsiou, D.; Granchi, Z.; Mitsiadis, T.A.; Jimenez-Rojo, L. Generation of Spheres from Dental Epithelial Stem Cells. Front. Physiol. 2017, 8, 7.
- Pagella, P.; Neto, E.; Lamghari, M.; Mitsiadis, T.A. Investigation of orofacial stem cell niches and their innervation through microfluidic devices. Eur. Cell. Mater. 2015, 29, 213–223.
- Pagella, P.; Cordiale, A.; Marconi, G.; Trubiani, O.; Rasponi, M.; Mitsiadis, T. Bioengineered tooth emulation systems for regenerative and pharmacological purposes. Eur. Cell. Mater. 2021, 41, 502–516.
- Vorwald, C.E.; Joshee, S.; Leach, J.K. Spatial localization of endothelial cells in heterotypic spheroids influences Notch signaling. J. Mol. Med. Berl. Ger. 2020, 98, 425–435.
- De Francesco, E.; Maggiolini, M.; Musti, A. Crosstalk between Notch, HIF-1α and GPER in Breast Cancer EMT. Int. J. Mol. Sci. 2018, 19, 2011.
- Wang, W.-M.; Zhao, Z.-L.; Ma, S.-R.; Yu, G.-T.; Liu, B.; Zhang, L.; Zhang, W.-F.; Kulkarni, A.B.; Sun, Z.-J.; Zhao, Y.-F. Epidermal Growth Factor Receptor Inhibition Reduces Angiogenesis via Hypoxia-Inducible Factor-1α and Notch1 in Head Neck Squamous Cell Carcinoma. PLoS ONE 2015, 10, e0119723.
- Drost, J.; Clevers, H. Organoids in cancer research. Nat. Rev. Cancer 2018, 18, 407–418.
- Lancaster, M.A.; Knoblich, J.A. Organogenesis in a dish: Modeling development and disease using organoid technologies. Science 2014, 345, 1247125.
- Hofer, M.; Lutolf, M.P. Engineering organoids. Nat. Rev. Mater. 2021, 6, 402–420.
- Kim, J.; Koo, B.-K.; Knoblich, J.A. Human organoids: Model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 2020, 21, 571–584.
- Simian, M.; Bissell, M.J. Organoids: A historical perspective of thinking in three dimensions. J. Cell Biol. 2017, 216, 31–40.
- Mariani, J.; Vaccarino, F.M. Breakthrough Moments: Yoshiki Sasai’s Discoveries in the Third Dimension. Cell Stem Cell 2019, 24, 837–838.
- Corrò, C.; Novellasdemunt, L.; Li, V.S.W. A brief history of organoids. Am. J. Physiol. Cell Physiol. 2020, 319, C151–C165.
- De Lau, W.; Barker, N.; Low, T.Y.; Koo, B.-K.; Li, V.S.W.; Teunissen, H.; Kujala, P.; Haegebarth, A.; Peters, P.J.; van de Wetering, M.; et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature 2011, 476, 293–297.
- Sato, T.; Vries, R.G.; Snippert, H.J.; van de Wetering, M.; Barker, N.; Stange, D.E.; van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009, 459, 262–265.
- Sahu, S.; Sharan, S.K. Translating Embryogenesis to Generate Organoids: Novel Approaches to Personalized Medicine. iScience 2020, 23, 101485.
- Trujillo, C.A.; Gao, R.; Negraes, P.D.; Gu, J.; Buchanan, J.; Preissl, S.; Wang, A.; Wu, W.; Haddad, G.G.; Chaim, I.A.; et al. Complex Oscillatory Waves Emerging from Cortical Organoids Model Early Human Brain Network Development. Cell Stem Cell 2019, 25, 558–569.e7.
- Shimojo, H.; Ohtsuka, T.; Kageyama, R. Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron 2008, 58, 52–64.
- Shimojo, H.; Isomura, A.; Ohtsuka, T.; Kori, H.; Miyachi, H.; Kageyama, R. Oscillatory control of Delta-like1 in cell interactions regulates dynamic gene expression and tissue morphogenesis. Genes Dev. 2016, 30, 102–116.
- Kobayashi, T.; Mizuno, H.; Imayoshi, I.; Furusawa, C.; Shirahige, K.; Kageyama, R. The cyclic gene Hes1 contributes to diverse differentiation responses of embryonic stem cells. Genes Dev. 2009, 23, 1870–1875.
- Das, S.; Gordián-Vélez, W.J.; Ledebur, H.C.; Mourkioti, F.; Rompolas, P.; Chen, H.I.; Serruya, M.D.; Cullen, D.K. Innervation: The missing link for biofabricated tissues and organs. NPJ Regen. Med. 2020, 5, 11.
- Zhang, Y.S.; Aleman, J.; Shin, S.R.; Kilic, T.; Kim, D.; Mousavi Shaegh, S.A.; Massa, S.; Riahi, R.; Chae, S.; Hu, N.; et al. Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proc. Natl. Acad. Sci. USA 2017, 114, E2293–E2302.
- Wörsdörfer, P.; Takashi, I.; Asahina, I.; Sumita, Y.; Ergün, S. Do not keep it simple: Recent advances in the generation of complex organoids. J. Neural Transm. 2020, 127, 1569–1577.
- Zhang, C.; Zhao, Z.; Abdul Rahim, N.A.; van Noort, D.; Yu, H. Towards a human-on-chip: Culturing multiple cell types on a chip with compartmentalized microenvironments. Lab. Chip 2009, 9, 3185–3192.
- Cavo, M.; Fato, M.; Peñuela, L.; Beltrame, F.; Raiteri, R.; Scaglione, S. Microenvironment complexity and matrix stiffness regulate breast cancer cell activity in a 3D in vitro model. Sci. Rep. 2016, 6, 35367.
- Giobbe, G.G.; Crowley, C.; Luni, C.; Campinoti, S.; Khedr, M.; Kretzschmar, K.; De Santis, M.M.; Zambaiti, E.; Michielin, F.; Meran, L.; et al. Extracellular matrix hydrogel derived from decellularized tissues enables endodermal organoid culture. Nat. Commun. 2019, 10, 5658.
- Ng, S.; Tan, W.J.; Pek, M.M.X.; Tan, M.-H.; Kurisawa, M. Mechanically and chemically defined hydrogel matrices for patient-derived colorectal tumor organoid culture. Biomaterials 2019, 219, 119400.
- Bhatia, S.N.; Ingber, D.E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014, 32, 760–772.
- Low, L.A.; Mummery, C.; Berridge, B.R.; Austin, C.P.; Tagle, D.A. Organs-on-chips: Into the next decade. Nat. Rev. Drug Discov. 2020, 20, 345–361.
- Pagella, P.; Neto, E.; Jiménez-Rojo, L.; Lamghari, M.; Mitsiadis, T.A. Microfluidics co-culture systems for studying tooth innervation. Front. Physiol. 2014, 5, 326.
- Pagella, P.; Mitsiadis, T.A. Analysis of Tooth Innervation in Microfluidic Coculture Devices. Methods Mol. Biol. Clifton NJ 2020, 2155, 99–106.
- Ye, W.; Luo, C.; Li, C.; Huang, J.; Liu, F. Organoids to study immune functions, immunological diseases and immunotherapy. Cancer Lett. 2020, 477, 31–40.
- Votanopoulos, K.I.; Forsythe, S.; Sivakumar, H.; Mazzocchi, A.; Aleman, J.; Miller, L.; Levine, E.; Triozzi, P.; Skardal, A. Model of Patient-Specific Immune-Enhanced Organoids for Immunotherapy Screening: Feasibility Study. Ann. Surg. Oncol. 2020, 27, 1956–1967.
- Yuki, K.; Cheng, N.; Nakano, M.; Kuo, C.J. Organoid Models of Tumor Immunology. Trends Immunol. 2020, 41, 652–664.
- Radtke, F.; Fasnacht, N.; Macdonald, H.R. Notch signaling in the immune system. Immunity 2010, 32, 14–27.
- Polini, A.; del Mercato, L.L.; Barra, A.; Zhang, Y.S.; Calabi, F.; Gigli, G. Towards the development of human immune-system-on-a-chip platforms. Drug Discov. Today 2019, 24, 517–525.
- Ando, K.; Kanazawa, S.; Tetsuka, T.; Ohta, S.; Jiang, X.; Tada, T.; Kobayashi, M.; Matsui, N.; Okamoto, T. Induction of Notch signaling by tumor necrosis factor in rheumatoid synovial fibroblasts. Oncogene 2003, 22, 7796–7803.
- Palaga, T.; Buranaruk, C.; Rengpipat, S.; Fauq, A.H.; Golde, T.E.; Kaufmann, S.H.E.; Osborne, B.A. Notch signaling is activated by TLR stimulation and regulates macrophage functions. Eur. J. Immunol. 2008, 38, 174–183.
- Klinakis, A.; Lobry, C.; Abdel-Wahab, O.; Oh, P.; Haeno, H.; Buonamici, S.; van De Walle, I.; Cathelin, S.; Trimarchi, T.; Araldi, E.; et al. A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature 2011, 473, 230–233.
- Franklin, R.A.; Liao, W.; Sarkar, A.; Kim, M.V.; Bivona, M.R.; Liu, K.; Pamer, E.G.; Li, M.O. The cellular and molecular origin of tumor-associated macrophages. Science 2014, 344, 921–925.
- Jimenez-Rojo, L.; Pagella, P.; Harada, H.; Mitsiadis, T. Dental Epithelial Stem Cells as a Source for Mammary Gland Regeneration and Milk Producing Cells In Vivo. Cells 2019, 8, 1302.
- Han, J.; Menicanin, D.; Gronthos, S.; Bartold, P.M. Stem cells, tissue engineering and periodontal regeneration. Aust. Dent. J. 2014, 59 (Suppl. S1), 117–130.
- Charbord, P. Bone marrow mesenchymal stem cells: Historical overview and concepts. Hum. Gene Ther. 2010, 21, 1045–1056.
- Mitsiadis, T.A.; Orsini, G.; Jimenez-Rojo, L. Stem cell-based approaches in dentistry. Eur. Cell. Mater. 2015, 30, 248–257.
- Orsini, G.; Pagella, P.; Mitsiadis, T.A. Modern Trends in Dental Medicine: An Update for Internists. Am. J. Med. 2018, 131, 1425–1430.
- Trubiani, O.; Marconi, G.D.; Pierdomenico, S.D.; Piattelli, A.; Diomede, F.; Pizzicannella, J. Human Oral Stem Cells, Biomaterials and Extracellular Vesicles: A Promising Tool in Bone Tissue Repair. Int. J. Mol. Sci. 2019, 20, 4987.
- Diomede, F.; Marconi, G.D.; Fonticoli, L.; Pizzicanella, J.; Merciaro, I.; Bramanti, P.; Mazzon, E.; Trubiani, O. Functional Relationship between Osteogenesis and Angiogenesis in Tissue Regeneration. Int. J. Mol. Sci. 2020, 21, 3242.
- Park, Y.-J.; Cha, S.; Park, Y.-S. Regenerative Applications Using Tooth Derived Stem Cells in Other Than Tooth Regeneration: A Literature Review. Stem Cells Int. 2016, 2016, 9305986.
- Xiao, L.; Kumazawa, Y.; Okamura, H. Cell death, cavitation and spontaneous multi-differentiation of dental pulp stem cells-derived spheroids in vitro: A journey to survival and organogenesis. Biol. Cell 2014, 106, 405–419.
- Moritani, Y.; Usui, M.; Sano, K.; Nakazawa, K.; Hanatani, T.; Nakatomi, M.; Iwata, T.; Sato, T.; Ariyoshi, W.; Nishihara, T.; et al. Spheroid culture enhances osteogenic potential of periodontal ligament mesenchymal stem cells. J. Periodontal Res. 2018, 53, 870–882.
- Kim, H.-J.; Sung, I.-Y.; Cho, Y.-C.; Kang, M.-S.; Rho, G.-J.; Byun, J.-H.; Park, W.-U.; Son, M.-G.; Park, B.-W.; Lee, H.-J.; et al. Three-Dimensional Spheroid Formation of Cryopreserved Human Dental Follicle-Derived Stem Cells Enhances Pluripotency and Osteogenic Induction Properties. Tissue Eng. Regen. Med. 2019, 16, 513–523.
- Ryu, N.-E.; Lee, S.-H.; Park, H. Spheroid Culture System Methods and Applications for Mesenchymal Stem Cells. Cells 2019, 8, 1620.
- Sugimoto, S.; Sato, T. Establishment of 3D Intestinal Organoid Cultures from Intestinal Stem Cells. Methods Mol. Biol. 2017, 1612, 97–105.
- Jeon, S.; Lee, H.-S.; Lee, G.-Y.; Park, G.; Kim, T.-M.; Shin, J.; Lee, C.; Oh, I.-H. Shift of EMT gradient in 3D spheroid MSCs for activation of mesenchymal niche function. Sci. Rep. 2017, 7, 6859.
- Lukomska, B.; Stanaszek, L.; Zuba-Surma, E.; Legosz, P.; Sarzynska, S.; Drela, K. Challenges and Controversies in Human Mesenchymal Stem Cell Therapy. Stem Cells Int. 2019, 2019, 9628536.
- Petrenko, Y.; Syková, E.; Kubinová, Š. The therapeutic potential of three-dimensional multipotent mesenchymal stromal cell spheroids. Stem Cell Res. Ther. 2017, 8, 94.
- Ferraro, F.; Celso, C.L.; Scadden, D. Adult stem cells and their niches. Adv. Exp. Med. Biol. 2010, 695, 155–168.
- Bejoy, J.; Bijonowski, B.; Marzano, M.; Jeske, R.; Ma, T.; Li, Y. Wnt-Notch Signaling Interactions during Neural and Astroglial Patterning of Human Stem Cells. Tissue Eng. Part A 2020, 26, 419–431.
- Iyer, K.; Chen, Z.; Ganapa, T.; Wu, B.M.; Tawil, B.; Linsley, C.S. Keratinocyte Migration in a Three-Dimensional In Vitro Wound Healing Model Co-Cultured with Fibroblasts. Tissue Eng. Regen. Med. 2018, 15, 721–733.
- Dosh, R.H.; Jordan-Mahy, N.; Sammon, C.; Le Maitre, C.L. Long-term in vitro 3D hydrogel co-culture model of inflammatory bowel disease. Sci. Rep. 2019, 9, 1812.
- Sun, L.; Wang, X.; Kaplan, D.L. A 3D cartilage—Inflammatory cell culture system for the modeling of human osteoarthritis. Biomaterials 2011, 32, 5581–5589.
- Damerau, A.; Gaber, T. Modeling Rheumatoid Arthritis In Vitro: From Experimental Feasibility to Physiological Proximity. Int. J. Mol. Sci. 2020, 21, 7916.
- Lv, D.; Hu, Z.; Lu, L.; Lu, H.; Xu, X. Three-dimensional cell culture: A powerful tool in tumor research and drug discovery. Oncol. Lett. 2017, 14, 6999–7010.
- Zhang, C.; Yang, Z.; Dong, D.-L.; Jang, T.-S.; Knowles, J.C.; Kim, H.-W.; Jin, G.-Z.; Xuan, Y. 3D culture technologies of cancer stem cells: Promising ex vivo tumor models. J. Tissue Eng. 2020, 11, 2041731420933407.
- Sorg, H.; Tilkorn, D.J.; Hager, S.; Hauser, J.; Mirastschijski, U. Skin Wound Healing: An Update on the Current Knowledge and Concepts. Eur. Surg. Res. 2017, 58, 81–94.
- Park, I.-S.; Chung, P.-S.; Ahn, J.C. Enhancement of Ischemic Wound Healing by Spheroid Grafting of Human Adipose-Derived Stem Cells Treated with Low-Level Light Irradiation. PLoS ONE 2015, 10, e0122776.
- Kim, S.; Kim, E.M.; Yamamoto, M.; Park, H.; Shin, H. Engineering Multi-Cellular Spheroids for Tissue Engineering and Regenerative Medicine. Adv. Healthc. Mater. 2020, 9, 2000608.
- Chigurupati, S.; Arumugam, T.V.; Son, T.G.; Lathia, J.D.; Jameel, S.; Mughal, M.R.; Tang, S.-C.; Jo, D.-G.; Camandola, S.; Giunta, M.; et al. Involvement of Notch Signaling in Wound Healing. PLoS ONE 2007, 2, e1167.
- Okuyama, R.; Nguyen, B.-C.; Talora, C.; Ogawa, E.; di Vignano, A.T.; Lioumi, M.; Chiorino, G.; Tagami, H.; Woo, M.; Dotto, G.P. High Commitment of Embryonic Keratinocytes to Terminal Differentiation through a Notch1-caspase 3 Regulatory Mechanism. Dev. Cell 2004, 6, 551–562.
- Del Campo, J.A.; Gallego, P.; Grande, L. Role of inflammatory response in liver diseases: Therapeutic strategies. World J. Hepatol. 2018, 10, 1–7.
- Tilg, H.; Diehl, A.M. Cytokines in Alcoholic and Nonalcoholic Steatohepatitis. N. Engl. J. Med. 2000, 343, 1467–1476.
- Ruoß, M.; Vosough, M.; Königsrainer, A.; Nadalin, S.; Wagner, S.; Sajadian, S.; Huber, D.; Heydari, Z.; Ehnert, S.; Hengstler, J.G.; et al. Towards improved hepatocyte cultures: Progress and limitations. Food Chem. Toxicol. 2020, 138, 111188.
- Nuciforo, S.; Heim, M.H. Organoids to model liver disease. JHEP Rep. 2021, 3, 100198.
- Huch, M.; Dorrell, C.; Boj, S.F.; van Es, J.H.; Li, V.S.W.; van de Wetering, M.; Sato, T.; Hamer, K.; Sasaki, N.; Finegold, M.J.; et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature 2013, 494, 247–250.
- Hu, H.; Gehart, H.; Artegiani, B.; LÖpez-Iglesias, C.; Dekkers, F.; Basak, O.; van Es, J.; Chuva de Sousa Lopes, S.M.; Begthel, H.; Korving, J.; et al. Long-Term Expansion of Functional Mouse and Human Hepatocytes as 3D Organoids. Cell 2018, 175, 1591–1606.e19.
- Kozyra, M.; Johansson, I.; Nordling, Å.; Ullah, S.; Lauschke, V.M.; Ingelman-Sundberg, M. Human hepatic 3D spheroids as a model for steatosis and insulin resistance. Sci. Rep. 2018, 8, 14297.
- Li, L.; Krantz, I.D.; Deng, Y.; Genin, A.; Banta, A.B.; Collins, C.C.; Qi, M.; Trask, B.J.; Kuo, W.L.; Cochran, J.; et al. Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat. Genet. 1997, 16, 243–251.
- Oda, T.; Elkahloun, A.G.; Pike, B.L.; Okajima, K.; Krantz, I.D.; Genin, A.; Piccoli, D.A.; Meltzer, P.S.; Spinner, N.B.; Collins, F.S.; et al. Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nat. Genet. 1997, 16, 235–242.
- Guan, Y.; Xu, D.; Garfin, P.M.; Ehmer, U.; Hurwitz, M.; Enns, G.; Michie, S.; Wu, M.; Zheng, M.; Nishimura, T.; et al. Human hepatic organoids for the analysis of human genetic diseases. JCI Insight 2017, 2, e94954.
- Spill, F.; Reynolds, D.S.; Kamm, R.D.; Zaman, M.H. Impact of the physical microenvironment on tumor progression and metastasis. Curr. Opin. Biotechnol. 2016, 40, 41–48.
- Tevis, K.M.; Colson, Y.L.; Grinstaff, M.W. Embedded Spheroids as Models of the Cancer Microenvironment. Adv. Biosyst. 2017, 1, 1700083.
- Tamaki, M.; McDonald, W.; Amberger, V.R.; Moore, E.; Del Maestro, R.F. Implantation of C6 astrocytoma spheroid into collagen type I gels: Invasive, proliferative, and enzymatic characterizations. J. Neurosurg. 1997, 87, 602–609.
- Porcheri, C.; Meisel, C.T.; Mitsiadis, T.A. Molecular and Cellular Modelling of Salivary Gland Tumors Open New Landscapes in Diagnosis and Treatment. Cancers 2020, 12, E3107.
- Zanoni, M.; Cortesi, M.; Zamagni, A.; Arienti, C.; Pignatta, S.; Tesei, A. Modeling neoplastic disease with spheroids and organoids. J. Hematol. Oncol. 2020, 13, 97.
- Weiswald, L.-B.; Bellet, D.; Dangles-Marie, V. Spherical cancer models in tumor biology. Neoplasia 2015, 17, 1–15.
- Hellström, M.; Phng, L.-K.; Hofmann, J.J.; Wallgard, E.; Coultas, L.; Lindblom, P.; Alva, J.; Nilsson, A.-K.; Karlsson, L.; Gaiano, N.; et al. Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature 2007, 445, 776–780.
- Siebel, C.; Lendahl, U. Notch Signaling in Development, Tissue Homeostasis, and Disease. Physiol. Rev. 2017, 97, 1235–1294.
- Lobry, C.; Oh, P.; Aifantis, I. Oncogenic and tumor suppressor functions of Notch in cancer: It’s NOTCH what you think. J. Exp. Med. 2011, 208, 1931–1935.
- Ayla, S.; Bilir, A.; Soner, B.C.; Yilmaz-Dilsiz, O.; Ergüven, M.; Oktem, G. Notch signaling-related therapeutic strategies with novel drugs in neuroblastoma spheroids. J. Pediatr. Hematol. Oncol. 2014, 36, 37–44.
- Xiao, Y.; Ye, Y.; Zou, X.; Jones, S.; Yearsley, K.; Shetuni, B.; Tellez, J.; Barsky, S.H. The lymphovascular embolus of inflammatory breast cancer exhibits a Notch 3 addiction. Oncogene 2011, 30, 287–300.
- Rebay, I.; Fleming, R.J.; Fehon, R.G.; Cherbas, L.; Cherbas, P.; Artavanis-Tsakonas, S. Specific EGF repeats of Notch mediate interactions with Delta and serrate: Implications for notch as a multifunctional receptor. Cell 1991, 67, 687–699.
- Rebay, I.; Fehon, R.G.; Artavanis-Tsakonas, S. Specific truncations of Drosophila Notch define dominant activated and dominant negative forms of the receptor. Cell 1993, 74, 319–329.
- Miyaki, M.; Yamaguchi, T.; Iijima, T.; Takahashi, K.; Matsumoto, H.; Mori, T. Somatic Mutations of the CDC4 (FBXW7) Gene in Hereditary Colorectal Tumors. Oncology 2009, 76, 430–434.
- Babaei-Jadidi, R.; Li, N.; Saadeddin, A.; Spencer-Dene, B.; Jandke, A.; Muhammad, B.; Ibrahim, E.E.; Muraleedharan, R.; Abuzinadah, M.; Davis, H.; et al. FBXW7 influences murine intestinal homeostasis and cancer, targeting Notch, Jun, and DEK for degradation. J. Exp. Med. 2011, 208, 295–312.
- Camps, J.; Pitt, J.J.; Emons, G.; Hummon, A.B.; Case, C.M.; Grade, M.; Jones, T.L.; Nguyen, Q.T.; Ghadimi, B.M.; Beissbarth, T.; et al. Genetic Amplification of the NOTCH Modulator LNX2 Upregulates the WNT/β-Catenin Pathway in Colorectal Cancer. Cancer Res. 2013, 73, 2003–2013.
- Sancho, R.; Jandke, A.; Davis, H.; Diefenbacher, M.E.; Tomlinson, I.; Behrens, A. F-box and WD Repeat Domain-Containing 7 Regulates Intestinal Cell Lineage Commitment and Is a Haploinsufficient Tumor Suppressor. Gastroenterology 2010, 139, 929–941.
- Zhu, J.; Wang, J.; Shi, Z.; Franklin, J.L.; Deane, N.G.; Coffey, R.J.; Beauchamp, R.D.; Zhang, B. Deciphering Genomic Alterations in Colorectal Cancer through Transcriptional Subtype-Based Network Analysis. PLoS ONE 2013, 8, e79282.
- Lee, S.H.; Choi, N.; Sung, J.H. Pharmacokinetic and pharmacodynamic insights from microfluidic intestine-on-a-chip models. Expert Opin. Drug Metab. Toxicol. 2019, 15, 1005–1019.
- Candini, O.; Grisendi, G.; Foppiani, E.M.; Brogli, M.; Aramini, B.; Masciale, V.; Spano, C.; Petrachi, T.; Veronesi, E.; Conte, P.; et al. Author Correction: A Novel 3D In Vitro Platform for Pre-Clinical Investigations in Drug Testing, Gene Therapy, and Immuno-oncology. Sci. Rep. 2020, 10, 1845.
- Fatehullah, A.; Tan, S.H.; Barker, N. Organoids as an in vitro model of human development and disease. Nat. Cell Biol. 2016, 18, 246–254.
- Sung, J.H. Multi-organ-on-a-chip for pharmacokinetics and toxicokinetic study of drugs. Expert Opin. Drug Metab. Toxicol. 2021, 17, 969–986.
- Langhans, S.A. Three-Dimensional in Vitro Cell Culture Models in Drug Discovery and Drug Repositioning. Front. Pharmacol. 2018, 9, 6.
- Polini, A.; Prodanov, L.; Bhise, N.S.; Manoharan, V.; Dokmeci, M.R.; Khademhosseini, A. Organs-on-a-chip: A new tool for drug discovery. Expert Opin. Drug Discov. 2014, 9, 335–352.
- Gupta, N.; Liu, J.R.; Patel, B.; Solomon, D.E.; Vaidya, B.; Gupta, V. Microfluidics-based 3D cell culture models: Utility in novel drug discovery and delivery research. Bioeng. Transl. Med. 2016, 1, 63–81.
- Esch, E.W.; Bahinski, A.; Huh, D. Organs-on-chips at the frontiers of drug discovery. Nat. Rev. Drug Discov. 2015, 14, 248–260.
- Van Zundert, I.; Fortuni, B.; Rocha, S. From 2D to 3D Cancer Cell Models-The Enigmas of Drug Delivery Research. Nanomaterials 2020, 10, 2236.
- Lim, J.; Ching, H.; Yoon, J.-K.; Jeon, N.L.; Kim, Y. Microvascularized tumor organoids-on-chips: Advancing preclinical drug screening with pathophysiological relevance. Nano Converg. 2021, 8, 12.
- Prantil-Baun, R.; Novak, R.; Das, D.; Somayaji, M.R.; Przekwas, A.; Ingber, D.E. Physiologically Based Pharmacokinetic and Pharmacodynamic Analysis Enabled by Microfluidically Linked Organs-on-Chips. Annu. Rev. Pharmacol. Toxicol. 2018, 58, 37–64.
- Kimura, H.; Sakai, Y.; Fujii, T. Organ/body-on-a-chip based on microfluidic technology for drug discovery. Drug Metab. Pharmacokinet. 2018, 33, 43–48.
- Golde, T.E.; Koo, E.H.; Felsenstein, K.M.; Osborne, B.A.; Miele, L. γ-Secretase inhibitors and modulators. Biochim. Biophys. Acta BBA—Biomembr. 2013, 1828, 2898–2907.
- Andersson, E.R.; Lendahl, U. Therapeutic modulation of Notch signaling—Are we there yet? Nat. Rev. Drug Discov. 2014, 13, 357–378.
- Real, P.J.; Tosello, V.; Palomero, T.; Castillo, M.; Hernando, E.; de Stanchina, E.; Sulis, M.L.; Barnes, K.; Sawai, C.; Homminga, I.; et al. γ-secretase inhibitors reverse glucocorticoid resistance in T cell acute lymphoblastic leukemia. Nat. Med. 2009, 15, 50–58.
