IQ motif-containing GTPase-activating proteins (IQGAPs) modulate a wide range of cellular processes by acting as scaffolds and driving protein components into distinct signaling networks. Their functional states have been proposed to be controlled by members of the RHO family of GTPases, among other regulators. In this study, we show that IQGAP1 and IQGAP2 can associate with CDC42 and RAC1-like proteins but not with RIF, RHOD, or RHO-like proteins, including RHOA. This seems to be based on the distribution of charged surface residues, which varies significantly among RHO GTPases despite their high sequence homology. Although effector proteins bind first to the highly flexible switch regions of RHO GTPases, additional contacts outside are required for effector activation. Sequence alignment and structural, mutational, and competitive biochemical analyses revealed that RHO GTPases possess paralog-specific residues outside the two highly conserved switch regions that essentially determine the selectivity of RHO GTPase binding to IQGAPs. Amino acid substitution of these specific residues in RHOA to the corresponding residues in RAC1 resulted in RHOA association with IQGAP1. Thus, electrostatics most likely plays a decisive role in these interactions
1. Introduction
IQ motif-containing GTPase-activating proteins (IQGAPs) belong to the class of multidomain scaffold proteins that play central roles in the assembly of protein complexes and signaling networks
[1][2][3][4][5][6][7]. In humans, three IQGAP paralogs have been described. The ubiquitously expressed IQGAP1 is the best-characterized paralog. IQGAP2 is mostly expressed in the liver, prostate, kidney, thyroid, stomach, testis, platelets, and salivary glands, while IQGAP3 is found in the brain, lung, testis, and intestine
[8]. Multiple domains enable IQGAPs to interact with a large number of proteins and to modulate the spatiotemporal distributions of distinct signal-transducing protein complexes, including B/CRAF-MEK1/2-ERK1/2
[9][10][11], FGFR1-CDC42-NWASP-ARP2/3-actin
[12][13][14], TIAM1-RAC1-PAK6
[15][16], and CDC42/RAC1/CLIP170
[17][18]. IQGAP paralogs share similar domain organization and high sequence homology (
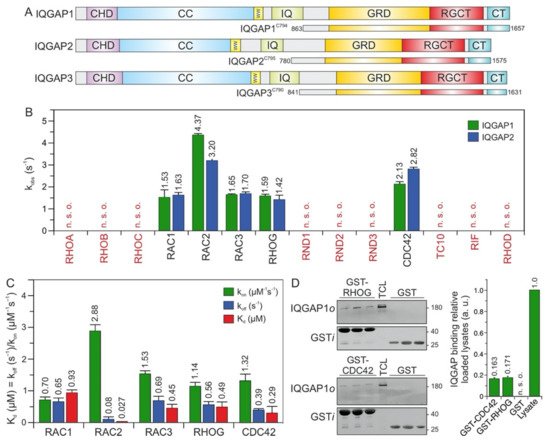
Figure 1A). The N-terminal calponin homology domain (CHD) binds F-actin
[19]. The polyproline-binding region (WW) binds ERK1/2
[9]. The IQ motif (IQ) binds HER1/2, KRAS, B/CRAF, MEK1/2, and calmodulin
[4][20][21][22][23][24]. The RASGAP-related domain (GRD) and RASGAP C-terminal domain (RGCT) bind to CDC42 and RAC1. The C-terminal domain (CT) binds E-cadherin, β-catenin, APC, and CLIP170
[3].
Figure 1. IQGAP1 and IQGAP2 selectively associate with CDC42 and RAC1-like proteins. (A) Domain organization of the IQGAP paralogs and their C-terminal fragments assessed in this study (see text for more details). (B) The association of IQGAP1C794 and IQGAP2C795 (2 µM) with various mGppNHp-bound RHO GTPases (0.2 µM). The kobs values for the interaction of IQGAP1 and IQGAP2 with several RHO GTPases, shown as bars, illustrate that both IQGAPs associate with CDC42 and RAC1-like proteins. The RHO-like proteins RND1, RND2, RND3, TC10, RIF, and RHOD did not associate with these IQGAPs under these conditions. (C) The association rates (kon) were measured using 0.2 µM mGppNHp-bound RHO GTPases with increasing concentrations (2–8 µM) of IQGAP1C794. Dissociation rates (koff) were measured by mixing 2 µM IQGAP1C794 complexed with mGppNHp-bound RHO GTPases (0.2 µM) and unlabeled RAC1-GppNHp (10 µM). The individual rate constants were calculated for the interaction of IQGAP1C794 with RAC- and CDC42-like proteins, and the results are plotted in bar charts. Association rates (kon), dissociation rates (koff), and dissociation constants (Kd) for IQGAP1C794-RHO protein binding are shown. RAC2 showed the highest binding affinity for IQGAP1C794, followed by CDC42, RAC3, RHOG, and RAC1. The data are expressed as the means ± S.D. All measurements were obtained in duplicate. n. s. o. = no signal observed. The data are expressed as the means ± S.D. (D) Binding of endogenous IQGAP1 to GppNHp-bound RHOG and CDC42 (left panel) was analyzed in a GST pull-down assay (n = 3) using total cell lysate (TCL) of HEK-293 cell (i, input; o, output). GST-CDC42•GppNHp was used as positive control. GST control experiments confirmed the specificity of the interaction between RHOG and IQGAP1. The upper part of the membrane was used for an anti-IQGAP1 immunoblotting, and the lower for an anti-GST. Densitometry analysis of relative IQGAP binding to GST-CDC42 or GST-RHOG (a. u., arbitrary unit) were performed in the next step. Bar charts at the right panel display the quantitation of detected signal in GST-pull down assay from a triplicate experiment.
CDC42 and RAC1 belong to the RHO GTPase family, which includes 20 classical paralogs
[25] that control diverse cellular functions
[26][27]. RHO GTPases are classified into six subfamilies: the RHO subfamily (RHOA, RHOB, and RHOC); the RAC subfamily (RAC1, RAC1B, RAC2, RAC3, and RHOG); the CDC42 subfamily (CDC42, G25K, TC10, TCL, WRCH1, and WRCH2); the RND subfamily (RND1, RND2, and RND3); and RHOD, RIF, and RHOH, which do not precisely fall into any of these subfamilies
[25][28]. The RHOBTB and MIRO subfamilies are atypical members of the RHO family that are structurally different from classical RHO family members and possess other additional functional domains
[29].
RHO GTPases are molecular switches that cycle between an inactive (GDP-bound) and an active (GTP-bound) form
[28]. In the active state, they interact with a multitude of target (effector) proteins, such as IQGAPs, to induce cellular responses
[30][31][32]. Interaction with RHO GTPases, such as CDC42 or RAC1, and/or phosphorylation of Ser-1441 and Ser-1443 may release IQGAPs from an autoinhibited state and induce their activated signaling competent state
[20][22][23][33]. The interaction of the C-terminal half of IQGAP1, encompassing the GRD, RGCT, and CT domains (hereafter called C794), with RAC1 and CDC42 has been intensively studied by several groups
[21][23][33][34][35][36][37][38][39]. Despite the common binding properties of CDC42 and RAC1 to IQGAPs, there are significant differences, which may be attributed to divergent IQGAP-RHO GTPase complexes that control distinct cellular processes
[23][24][36][40][41]. As the highly flexible switch I and II regions (encompassing amino acids 29–42 and 62–68, respectively), which change their conformation upon GDP to GTP exchange
[31], are almost identical in CDC42 and RAC1, the selectivity-determining residues need to be located outside these two regions.
2. IQGAP1/2 Selectively Bind CDC42 and RAC1-Like Members of the RHO Family
The C-terminal 794 amino acids (aa) of IQGAP1, encompassing the GRD, RGCT, and CT domains, and C795 of IQGAP2, were successfully purified to measure their binding properties over a broad range of RHO GTPases. Interaction studies were performed using time-resolved stopped-flow fluorescence (SFF) spectrometry under previously described conditions
[23]. Accordingly, both IQGAPs were associated similarly with the active forms of RAC1, RAC2, RAC3, RHOG, and CDC42, but not with RND1, RND2, RND3, TC10, RHOA, RHOB, RHOC, RHOD, or RIF (
Figure 1B).
RND proteins represent a distinct group of proteins within the RHO family. They were purified in their GTP-bound state, but replacing GTP with mGppNHp, 2′/3′-O-(N-methyl-anthraniloyl)- guanosine-5′-[(β,γ)-imido]triphosphate, a slow-hydrolyzing analog of GTP, resulted in complex instability. Therefore, for interaction studies with IQGAPs, we performed indirect competitive assays. We measured the association of IQGAP1C794 with RAC1 in the presence and absence of excess GTP-bound RND proteins. As a positive control, we used GppNHp-bound CDC42. In contrast to CDC42, which competitively blocked the IQGAP-RAC1 interaction, no binding of RND proteins was observed (Figure 1B), suggesting that IQGAP1 and IQGAP2 do not interact with these unconventional members of the RHO family.
Given these findings, it was important to investigate the complex formation and binding stoichiometry between CDC42/RAC1 and the IQGAP proteins. LeCour et al. have proposed that constitutively active CDC42
Q61L but not RAC1
Q61L binds the IQGAP2 (GAP)-related domain (GRD) in a 2:1 ratio to promote IQGAP2 dimerization
[41]. Therefore, we performed analytical size-exclusion chromatography using IQGAP1
C794 and IQGAP2
C795 alone or mixed with CDC42•GppNHp or RAC1•GppNHp. The elution profiles showed that CDC42, RAC1, and IQGAP1 eluted as dimers, while IQGAP2 eluted mainly as monomers and to some extent as trimers and tetramers. The elution profiles of the IQGAPs mixed with CDC42 and RAC1 showed, in addition to RAC1 and CDC42 (peak #1), two peaks (#3 and #5), indicating molecular weights (
MWs) of 222–235 kDa and elution volumes of 10.2–11.0 mL. Coomassie-brilliant-blue-stained SDS-PAGE gels revealed that only peak #3 contained IQGAP complexes with RAC1 and CDC42, with an average
MW of 228 kDa that corresponds to a heterotetramer. IQGAP2
C795 also eluted as higher oligomers (peak #6), which did not contain either RAC1 or CDC42.
LeCour et al. have reported a high affinity interaction between CDC42
Q61L and IQGAP GRD (41). In our previous study, we have shown that CDC42
Q61L has a 13-fold stronger interaction with GRD as compared with CDC42
WT (23). Therefore, we purified and investigated the stoichiometry of CDC42
Q61L•GppNHp for its complex formation with IQGAP1 GRD in direct comparison with CDC42
WT•GppNHp. In the case of CDC42
Q61L, the elution profile represented two peaks for the GRD and CDC42
Q61L complex, corresponding to heterotrimeric complex with a stoichiometry of 2:1, as proposed by LeCour et al.
[23][41]. However, GRD and CDC42
WT complex eluted as a heterotetramer.
Overall, the analyses of the size-exclusion chromatography data suggest that under our experimental conditions, the composition of the IQGAP1/2 complexes with both RAC1 and CDC42 corresponds to a 2:2 ratio. Furthermore, the CDC42-GppNHp elution profile at 15.6 and 15.9 mL of elution volume confirmed the previous observations reported by Zhang et al. regarding the reversible homodimerization of RHO family GTPases
[42].
3. RAC2 Exhibited the Highest Affinity for IQGAP1
To examine binding properties, the respective association rate constants (kon) and the dissociation rate constants (koff) were determined for the interaction of IQGAP1C794 with CDC42 and RAC1-like proteins under the aforementioned conditions (Figure 1C). The values are in a range similar to that of wild-type RAC1, RAC3, RHOG, and CDC42, with the exception of RAC2, which strikingly showed a Kd value of 27 nM, the highest affinity for IQGAP1C794 (Figure 1C). The rapid association and slow dissociation rates are remarkable, and suggest that the RAC2–IQGAP1 interaction remains stable for a long residence time.
The IQGAP1C794 binding of RHOG, in addition to its binding to the RAC and CDC42 proteins, prompted us to investigate the association of RHOG with endogenous IQGAP1 using purified GST-RHOG•GppNHp as bait in a pull-down assay. GST was used as the negative control, and GST-CDC42•GppNHp was used as the positive control. Quantification of the immunoblot analysis using specific antibodies against GST and IQGAP1 showed that cellular IQGAP1 bound RHOG as efficiently as it bound CDC42 (Figure 1D).
Next, we performed an in-depth investigation of the IQGAP1C794 interactions with RAC1 and CDC42, which are widely acknowledged to be IQGAP-binding partners.
4. Potential Hotspots for IQGAP Binding Appear Outside the Switch Regions
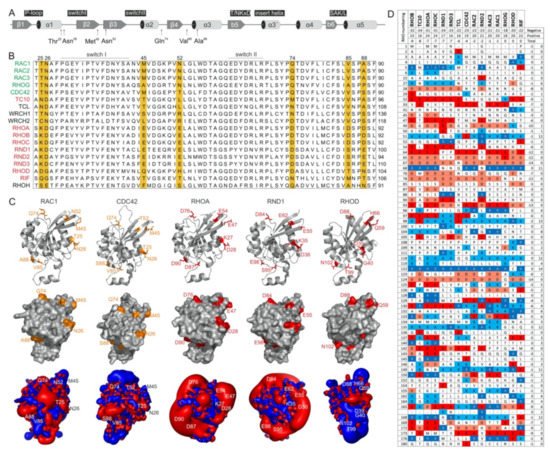
The switch regions (Figure 2A), which are generally known as effector binding sites, are required but not sufficient for effector binding selectivity. The amino acid sequences of these two regions are almost identical, which is particularly notable in comparison to IQGAP1-binding proteins (e.g., members of the RAC subfamily) with nonbinders (e.g., members of the RHO subfamily) (Figure 2B). Thus, a set of specificity-determining residues in RHO GTPase that direct interactions with IQGAPs must reside outside of the switch regions. In this context, notably, the CDC42 subfamily includes both IQGAP binders and nonbinders. Accordingly, we found four different hotspots, residues 25/26, 45/52, 74, and 85/88 (based on CDC42/RAC numbering), that are highly conserved in IQGAP1 binders and clearly deviate from the corresponding residues in nonbinders (Figure 2B). Notably, we did not consider residues that are quite variable not only between the binders and nonbinders but also within the IQGAP1 binders themselves (e.g., T24, A27, G30, S41; Figure 2B). Moreover, an inspection of the crystal structures of RHO GTPases in five different subfamilies showed that these four sites did not significantly contribute to local structural variations (Figure 2C, upper panel). These residues surround the switch regions and, most interestingly, are all located on the surface of the respective proteins and are thus available for intermolecular interactions (Figure 2C, middle panel).
Figure 2. RHO GTPases exhibit significantly different electrostatic properties. (A) The G domain organization of RAC1 indicates secondary structure elements, key functional regions and locations of residues crucial for IQGAP1 binding. (B) A multiple amino acid sequence alignment of canonical RHO GTPases revealed various residues outside of the switch regions that may determine their differential interactions with IQGAPs. IQGAP binders are colored green, and the nonbinders are colored red. (C) Structures in ribbon representation, solvent accessible proteins surfaces and electrostatic potential maps for RAC1 (PDB code, 1MH1), CDC42 (PDB code, 2QRZ), RHOA (PDB code, 1A2B), RND1 (PDB code, 2CLS), and RHOD (PDB code, 2J1L) are shown. Thr-25, Asn-26, Met-45, Asn-52, Gln-74, Val-85, and Ala-88 of RAC1 proposed to determine its specificity for the binding of IQGAPs are located on the surface, negatively charged residues on corresponding positions in, for example, RHOA and RND1 cause significant negative electrostatic potentials. Images were generated with the PyMOL molecular viewer. (D) The distribution of charged amino acids vary significantly among RHO GTPases despite their high sequence homology. Sequence alignment of the RHO GTPases used in this study reduced in a way that only loci containing at least one positively charged amino acid, i.e., arginine or lysine, or one negatively charged amino acid, i.e., glutamate or aspartate, were retain, respectively. It demonstrates diverse occurrence of charges in proteins molecules of RHO GTPases that is also reflected on huge differences of electrostatic potentials shown in C. They roughly also correspond to theoretical net charges for whole proteins that were obtained as sums of the +1 or −1 for positively or negatively charged residues, respectively. As only RHOoD and RIF were found to be electrically neutral while all other GTPases possess overall negative net charge, characteristic lobes of negative, red colored electrostatic potentials around the majority of proteins were observed.
As almost all amino acids at the selected hotspots in RHOA and RND proteins have charged side chains, electrostatics very likely play a crucial role in complex formation with IQGAP1. With the aim of verifying this hypothesis, we first calculated the electrostatic potentials around these molecules. While the form and magnitude of the electrostatic isosurfaces for cognate RHO GTPases were found to be similar, striking differences were found between their subclasses, with particularly strong negative potentials in the cases of TC10, TCL, RHOA, and RND proteins (Figure 2C, lower panel). Aiming to understand the origin of these differences in the electrostatic potentials of the 15 examined RHO GTPases, we calculated the net charges of their G-domains with −1 attributed to aspartic or glutamic acid and +1 attributed to arginine or lysine. Although RHO GTPases are highly homologous, variations in particular amino acids that might seem negligible from a sequence point of view can lead to a broad span of net protein charges. In the cases of the studied GTPases, the span of electrostatic charges ranges from −9 for RHOB to electrically neutral RIF, clearly explaining the differences in electrostatic potential. The larger the lobe of the negative potential around the protein is, the more negative its net charge. Correlating the electrostatic charge with the binding to IQGAP1, negative charges might discriminate the association with TC10, TCL, RHOA, and RND paralogs. On the other hand, balanced potentials seem to be just a prerequisite for binding, because the charges and corresponding electrostatic potentials of all other GTPases are similar, but RHOD and RIF belong to nonbinders.
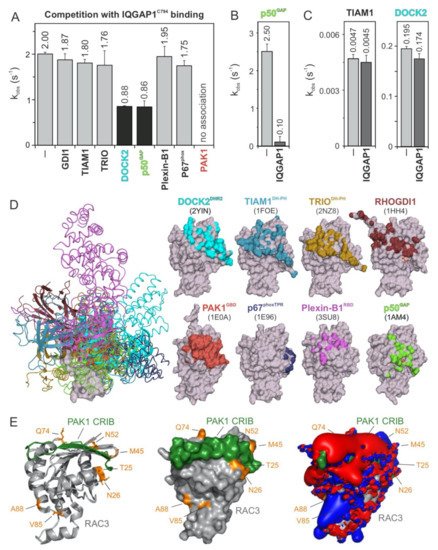
5. PAK1, p50GAP, and DOCK2 Compete with IQGAP1 for Binding RAC1
To further map the IQGAP1
C794-binding regions on the surface of the RAC1 structure, we performed competitive binding experiments. We repeated the measurement of the IQGAP1
C794 association with RAC1•mGppNHp in the absence and presence of a 10-fold molar excess of other RAC1-interacting proteins that may be competitors: full-length GDI1, the DBL homology-pleckstrin homology tandem (DH-PH) domain of TIAM1 and TRIO, the DOCK homology region 2 (DHR2) domain of DOCK2, the GAP domain of p50
GAP, the GTPase-binding domain (GBD) of PAK1, the RAC1-binding domain of plexin-B1 RBD, and the tetratricopeptide repeat (TPR) of p67
Phox (
Figure 3). These proteins were premixed with IQGAP1
C794 before rapid mixing with RAC1•mGppNHp in a stopped-flow apparatus. The working model was based on the presumption that if the binding of RAC1 to IQGAP1
C794 and to RAC1-interacting proteins is mutually exclusive, then the proteins in the mixture will interfere with the ability of IQGAP1
C794 to associate with RAC1. As shown in
Figure 3A, IQGAP1
C794 association with RAC1•GppNHp was partially abolished with DOCK2 and p50
GAP, completely abolished with PAK1, and not affected by the other proteins. Notably, GEFs, and most likely DOCK2, do not significantly distinguish between GDP- and GTP- (or GppNHp-) bound RHO GTPases
[43].
Figure 3. IQGAP1C794 competes with DOCK2, p50GAP, and PAK1 for binding RAC1. (A) The evaluated observed rate constants (kobs), shown as bars, demonstrate that IQGAP1C794 associates with RAC1 regardless of the presence of excess amounts of GDI1, TIAM1, TRIO, Plexin-B1, or p67phox, while the association was blocked in the presence of DOCK2 or p50GAP and completely abolished in the presence of PAK1. (B) The p50GAP-stimulated GTPase activity of RAC1 was drastically reduced in the presence of IQGAP1C794. (C) TIAM1- and DOCK2-catalyzed nucleotide exchange activity of RAC1 was not significantly changed in the presence of excess amounts of IQGAP1C794. The data are expressed as the means ± S.D. (D) The left panel shows the structure of RAC1 (gray represents the surface) in complex with different RAC and CDC42 interacting partners (in different colored ribbons), including DOCK2DHR2, TIAM1DH-PH, TRIODH-PH, p50GAP, GDI1, PAK1GBD, p67phoxTRP, and Plexin-B1RBD. The right panel highlights the contact sites of these binding proteins on the surface of RAC1 in the corresponding colors. The protein database identification codes of the respective structures are indicated. (E) The complex structure of RAC3 (PDB code, 2IC5) and the CRIB motif of PAK1 (PDB code, 2QME) shows that T25, N26, M45, N52, and Q74 of RAC3 are in close proximity to the CRIB motif-binding region. Electrostatic potentials (right panel) show that the PAK1 CRIB motif generates an overall negative electrostatic surface potential.
In addition, we measured the impact of IQGAP1
C794 binding to RAC1 on the GEF and GAP activities of TIAM1, DOCK2, and p50
GAP (
Figure 3B,C). The speculation that GEFs may compete with IQGAP1
C794 for RAC1•GDP binding is based on the assumption that IQGAP1
C794 binds to other sites outside the switch regions
[40]. No change was observed for the nucleotide exchange reaction catalyzed by TIAM1 or DOCK2 (
Figure 3C), corroborating our previous observation that IQGAP1
C794, which binds CDC42•GDP, does interact with RAC1•GDP
[23]. In contrast, p50
GAP-stimulated GTP hydrolysis activity was drastically inhibited, reduced by 25-fold, in the presence of IQGAP1
C794 (
Figure 3B), confirming the selective and high-affinity binding of IQGAP1
C794 to RAC1•GTP.
To determine which amino acids of RAC1 are critical for the observed interactions and effects, we first overlaid the extracted structures of the investigated binding proteins (Figure 3D, left panel) with residues that form the interacting interfaces and depicted them on a surface representation of the respective RAC1 structures (Figure 3D, right panel). The interacting interfaces are shown in colors corresponding to the RAC1-binding proteins. We further analyzed the crystal structure of RAC3 in complex with PAK1 GBD, which fully interfered with IQGAP1C794 binding to RAC1, and may thus share overlapping binding regions. Remarkably, the residues previously identified by sequence structural analysis as potential (hot)spots for the association of RHO GTPases with IQGAP1C794, namely, T25, N26, M45, N52, and Q74 of RAC3, are located in proximity of the RAC1-binding region of PAK1 GBD (Figure 3E). Visualizing the electrostatic potential of this complex structure showed that PAK1 GBD generates an overall negative electrostatic potential on the surface of RAC3 (Figure 3E, right panel).
6. IQGAP1 Binding Hotspots Significantly Vary among RHO GTPases
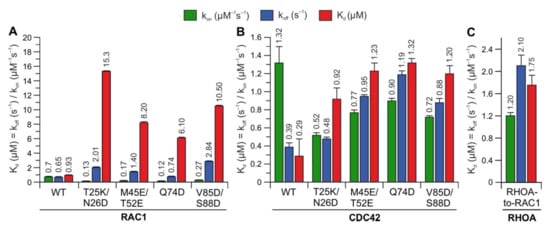
To identify whether the predicted hotspots determine differences in the interaction of IQGAP1
C794 with RHO proteins, we replaced these sites in RAC1 and CDC42 with the corresponding amino acids in RHOA (T25K/N26D, M45E/N52E, Q74D, and V85D/S88D) (
Figure 2A). Notably, S88 of CDC42 is in the same region as IQGAP2-contacting residues
[41]. The interaction of these variants with IQGAP1
C794 was measured under the same conditions as described above. Strikingly, major changes in the IQGAP1
C794 binding kinetics were observed for the RAC1 variants but not for the CDC42 variants (
Figure 4, left and middle panels). All the variants exhibited slower association kinetics and faster dissociation kinetics. As a result, the overall decrease in the binding affinities of the RAC1 variants for IQGAP1
C794 ranged between 7- and 17-fold, suggesting that these residues are either part of the RAC1–IQGAP1 binding interface or in close proximity to the IQGAP1-binding sites. To identify the impact of these residues, we generated a RHOA variant containing five substitutions, K27T, D28N, E47M, E54N, and D76Q, to mimic RAC1. Interestingly, this RHOA-to-RAC1 variant was capable of associating with IQGAP1
C794, while RHOA
WT did not show any association with IQGAP1
C794 (
Figure 4, right panel). These data confirmed the identified sequence-specific binding sites as hotspots.
Figure 4. Kinetic measurements of RAC1 and CDC42 variants binding IQGAP1C794. The calculated association rates (kon), dissociation rates (koff), and dissociation constants (Kd) for the interaction of IQGAP1C794 with different variants of RAC1 (A), CDC42 (B), and RHOA (C) are plotted as bar charts.
This entry is adapted from the peer-reviewed paper 10.3390/ijms222212596