Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Biochemistry & Molecular Biology
Le proteine VDAC (canale selettivo anionico voltaggio-dipendente), note anche come suine mitocondriali, sono le proteine più abbondanti della membrana mitocondriale esterna (OMM), dove svolgono un ruolo vitale in vari processi cellulari, nella regolazione del metabolismo e nelle vie di sopravvivenza.
- voltage dependent anion channel
- cysteine overoxidation
- deamidation
1. Introduzione
1.1. Isoforme VDAC: una famiglia di proteine hub
Le proteine VDAC (canale selettivo anionico voltaggio-dipendente), note anche come suirine mitocondriali, sono le proteine più abbondanti della membrana mitocondriale esterna (OMM) dove svolgono un ruolo vitale in vari processi cellulari, nella regolazione del metabolismo e nelle vie di sopravvivenza. Mediano lo scambio di ioni e metaboliti tra i mitocondri e il resto della cellula, garantendo una buona funzionalità dei complessi mitocondriali e la produzione di energia [1].
Negli eucarioti superiori, ci sono tre isoforme VDAC (VDAC1, VDAC2, VDAC3) codificate da geni separati situati su cromosomi diversi [2]. Queste proteine che formano i pori hanno un peso molecolare simile (30 kDa) e sequenze altamente conservate di circa 280 amminoacidi ad eccezione del VDAC2 che ha la frazione N-terminale di 11 residui più lunga delle altre isoforme.
L'analisi evolutiva indica VDAC3 come l'isoforma più antica, mentre VDAC1 è considerata la più giovane porina mitocondriale [3][4].
Le strutture sperimentali 3D dell'isoforma VDAC1 di topo e umana sono state determinate utilizzando la cristallografia a raggi X e la NMR [5][6][7]. Queste analisi hanno rivelato una struttura costituita da 19 β-filamenti disposti a formare una trans-membrana β-barile e da una regione contenente α-elica al N-terminus della proteina. La canna è organizzata come una normale schiera antiparallela di β fili ad eccezione dei fili 1 e 19 che corrono in parallelo. La coda anfipatica a α elica si trova all'interno del poro. Tuttavia, la posizione esatta e la struttura locale di questo segmento sono ancora sfuggenti poiché queste caratteristiche non si sovrappongono perfettamente nelle strutture a raggi X e NMR disponibili [5][6][7].
Recentemente, la struttura di zebrafish VDAC2 è stata risolta ad alta risoluzione, confermando la stessa disposizione β-barrel di VDAC1 [8]. Zebrafish VDAC2 ha un residuo di cisteina nella sequenza e manca della sequenza N-terminale più lunga di 11 amminoacidi presente nelle VDAC dei mammiferi.
La struttura VDAC3 non è stata ancora determinata. Diverse previsioni bioinformatiche, basate sulla somiglianza di grandi sequenze, hanno proposto un nucleo di botte come le altre isoforme VDAC [9]. Nonostante l'elevata somiglianza di sequenza e l'omologia strutturale, le isoforme VDAC mostrano diverse proprietà funzionali all'interno della cellula.
L'analisi dei livelli di espressione delle isoforme VDAC umane nelle cellule HeLa, determinati mediante REAL-TIME PCR, suggerisce che VDAC1 è l'isoforma più abbondante, dieci volte più abbondante rispetto a VDAC2 e cento volte più abbondante rispetto a VDAC3, la meno caratterizzata delle isoforme. Inoltre, la sovraespressione di ogni singola isoforma VDAC influisce sui livelli di mRNA delle altre due isoforme, suggerendo che i rapporti tra le isoforme VDAC sono sottoposti a un controllo reciproco che evita uno squilibrio tra queste proteine [10].
Sebbene le tre isoforme mostrino un coinvolgimento comune nel mantenimento della bioenergetica cellulare, VDAC1 e VDAC2 hanno funzioni specializzate nella morte cellulare programmata. Per l'isoforma VDAC3, studi recenti indicano un ruolo centrale nel metabolismo dei ROS e nel controllo di qualità mitocondriale [11].
Le funzioni dei VDC sono parecchie e alcune di queste dipendono o sono influenzate dall'interazione con altre proteine citosoliche e mitocondriali. A causa della loro localizzazione presso l'OMM, le VDAC sono considerate proteine hub, che interagiscono con oltre 200 proteine al fine di integrare le funzioni mitocondriali con il resto delle attività cellulari [12][13]. Pertanto, le isoforme VDAC sembrano essere una giunzione per una varietà di segnali associati a diversi percorsi legati alla sopravvivenza cellulare o alla morte programmata. Inoltre, la funzione dei VDAC e le loro interazioni con altre proteine sono influenzate da modificazioni post-traduzionali (PTM) [14]. Sfortunatamente, i PTM delle proteine VDAC rappresentano un campo poco esplorato, principalmente perché la scoperta e la caratterizzazione del PTM in queste proteine è molto impegnativa, a causa della loro scarsa solubilità e impossibilità di isolare singole isoforme. Solo negli ultimi anni, il crescente numero di strumenti volti a identificare e quantificare i PTM ha migliorato le conoscenze in questo campo e nei meccanismi che regolano le funzioni e le interazioni delle suipine mitocondriali. In particolare, lo sviluppo di metodi di cromatografia liquida ad altissime prestazioni (nanoRP-UHPLC) e di spettrometria di massa ad alta risoluzione (HRMS) a fase nano-invertita ha svolto un ruolo chiave in questo campo. I risultati ottenuti sui PTM VDAC utilizzando tali metodologie, che hanno permesso una caratterizzazione approfondita di queste proteine dei pori trans-membrana molto idrofobiche, sono riassunti in questa recensione.
1.2. Le VDAC come attori principali nella mediazione e nella regolazione delle funzioni mitocondriali con attività cellulari
La posizione nell'OMM consente alle proteine VDAC di agire come punti di ancoraggio per diversi insiemi di molecole che interagiscono con i mitocondri. In questo modo, i VDC sono in grado di mediare e regolare l'integrazione delle funzioni mitocondriali con le attività cellulari.
L'interattoma VDAC comprende proteine situate in OMM, membrana mitocondriale interna (IMM), spazio intermembrana (IMS), citosol, reticolo endoplasmatico, membrana plasmatica e nucleo che sono coinvolti nel metabolismo, apoptosi, trasduzione del segnale, protezione contro ROS, legame a RNA o DNA e altro ancora.
La mobilità della regione VDAC N-terminale α-elica è importante per il gating dei canali, ma anche per le interazioni con proteine pro-apoptotiche e anti-apoptotiche come Bax, Bak e Bcl-xL [15][16][17]. Il VDAC1 è coinvolto nel rilascio di fattori apoptotici localizzati nello spazio intermembrana grazie alla sua capacità di oligomerizzarsi in dimeri, esameri e strutture di ordine superiore, per formare un grande poro che consente il passaggio del citocromo c e del fattore di induzione dell'apoptosi (AIF) al citosol e di conseguenza l'attivazione della morte cellulare programmata. Invece, VDAC2 funziona come fattore anti-apoptotico ed è sovraregolato in diverse malattie debilitanti tra cui l'Alzheimer e il cancro [18]. Questa proprietà è probabilmente dovuta alla capacità unica di VDAC2 di sequestrare la proteina pro-apoptotica Bak nell'OMM e mantenerla nello stato inattivo [11].
VDAC1 mostra siti di legame, situati nella sua porzione citosolica, per molti enzimi metabolici, come la gliceraldeide-3-fosfato deidrogenasi, la creatinchinasi, la glicerolo chinasi, la glucochinasi, la chinasi c-Raf e le isoforme e l'esochinasi (I e II), che necessitano di un accesso preferenziale all'ATP mitocondriale [19].
L'esochinasi interagisce attraverso la sua sequenza idrofobica N-terminale con Glu73 di VDAC1, un sito di legame localizzato su un lato della parete della canna, sepolto nell'ambiente idrofobo di OMM [20].
È stato dimostrato che il trattamento dei mitocondri con dicicloesilcarbodiimide (DCCD) inibisce l'interazione esochinasi-VDAC a causa della modifica chimica selettiva della glu73 [21].
Il residuo di glu73 è anche il sito di legame per le ceramidi, lipidi oncosoppressori in grado di agire direttamente sui mitocondri per innescare la morte delle cellule apoptotiche. È interessante notare che sia VDAC1 che 2 possiedono, in una posizione simile, un residuo di cisteina (Cys127 nel VDAC1 umano e Cys138 nel VDAC2 umano) sotto forma di acido solfonico con una forte carica negativa simile a quella del residuo acido glutammato [22]. Invece, l'isoforma VDAC3 non mostra alcun residuo omologo a Cys127/138 o Glu73 incorporato nella porzione idrofobica dell'OMM.
VDAC1 presenta una tasca legante il colesterolo formata, in isoforma umana, da residui di Ile123,Leu144,Tyr146,Ala151e Val171 [ 23].
Le suine mitocondriali formano complessi con altre proteine, come il nucleotide translocase dell'adenina (ANT), la proteina traslocatrice (TSPO), nota anche come recettore delle benzodiazepine di tipo periferico (PBR), HSP70 mitocondriale e diverse proteine citoscheletriche come tubulina, actina, catena leggera della dineina e gelsolina [11].
La proteina traslocatrice interagisce direttamente con tutte le isoforme VDAC. In particolare, l'interazione tra TSPO e VDAC1 contribuisce a regolare l'efficienza dei meccanismi di controllo della qualità mitocondriale e inibisce la mitofagia, prevenendo l'ubiquitinazione delle proteine attraverso la downregulation della via PINK1/Parkin [24]. Il motivo GxxxG si presenta sia in VDAC che in TSPO, ed è necessario per questa interazione [25]. Inoltre, VDAC1 e TSPO, in associazione con StAR (steroidogenic acute regulatory protein), formano il trasdusoma, un complesso multiproteico coinvolto nel trasporto del colesterolo. In una precedente ipotesi, VDAC1 e TSPO in OMM, ANT in IMM e Cyclophilin D nella matrice mitocondriale erano candidati a costituire il poro di transizione di permeabilità (PTP), un poro ad alta conduttanza e non specifico che consente il gonfiore mitocondriale e il rilascio di proteine apoptogeniche. Più recentemente, è stato proposto che il PTP potesse essere formato da dimeri del complesso ATP sintasi [26].
Recenti studi hanno focalizzato l'attenzione sul ruolo delle proteine VDAC nella disfunzione mitocondriale tipica di molte condizioni patologiche tra cui ictus, cancro, encefalomiopatie mitocondriali e invecchiamento, nonché disturbi neurodegenerativi [27][28].
L'analisi della spettrometria di massa ha rivelato l'associazione tra VDAC e l'ubiquitina ligasi Parkin. In presenza di mitocondri danneggiati, come nel morbo di Parkinson, La Parkina viene fosforilata da PINK1 e di conseguenza ubiquitina le proteine che risiedono sull'OMM, prendendo di mira i mitocondri per la degradazione. Parkin è una proteina citosolica ma trasloca nei mitocondri per partecipare ai meccanismi di controllo della qualità mitocondriale. Le proteine VDAC rappresentano un sito di attracco di Parkin sui mitocondri difettosi [29].
Moreover, VDAC1 represents the main docking site at the mitochondrial level for misfolded and aggregated proteins, a common feature of neurodegenerative disorders known as proteinopathies, such as Alzheimer’s disease (AD), Parkinson’s disease (PD), Creutzfeldt–Jacob disease (CJD), dementia with Lewy bodies (DLB), Huntington disease (HD), and amyotrophic lateral sclerosis (ALS) [30].
For example, in AD post-mortem brains, in neuroblastoma cells and in an AD mouse model, a direct association was demonstrated between VDAC1, specifically its N-terminal region, and hyper-phosphorylated Tau but also with amyloid beta (Aβ), both in its monomeric and oligomeric forms [31]. These interactions can have a dramatic effect on mitochondrial functions in AD neuron because they block the PTP formation, disrupt the transport of mitochondrial proteins and metabolites, and impair gating, conductance, and physiological interactome of VDACs [32].
In Parkinson’s disease, α-synuclein directly interacts with mitochondria, blocks VDAC1, and impairs metabolite fluxes leading, consequently, to an energetic crisis able to compromise cell viability [33].
In ALS, several SOD1 mutants are able to bind VDAC1 [34]. This interaction impairs ATP/ADP exchange, VDAC1 conductance and mitochondrial membrane potential. Recently, the competition between SOD1G93A and HK1 was demonstrated in binding VDAC1, in NSC34 motor-neuron cell lines [35].
In literature, the role of VDAC1 in neurodegeneration is rather well known; however, the involvement of the other two isoforms in these pathways remains poorly defined. This is likely associated with the relative abundance of VDAC1 compared to other isoforms which are more difficult to isolate in pure form.
Recent studies demonstrated that Cytoskeleton-associated protein 4 (CKAP4), a palmitoylated type II transmembrane protein localized to the endoplasmic reticulum (ER), regulates mitochondrial functions through an interaction with VDAC2 at ER-mitochondria contact sites [36].
VDAC2 binds inositol trisphosphate receptors (IP3R) and regulates the release of Ca2+ from the ER. In addition, several other interaction partners have been reported for VDAC2 isoform, which imply its effect in multiple cellular functions. Specifically, VDAC2 has been linked to many cellular proteins, including apoptotic factors as Bak and Bax, StAR, Metaxin2, eNOS (nitric oxide synthesize), GSK3β, tubulin, and Mcl1 [19]. In addition, VDAC2 and RACK1 (receptor of activated protein kinase C1) function as receptors for lymphocystis disease virus (LCDV) and for bursal disease virus in host cells [37].
VDAC2 together with VDAC3 binds Erastin, the activator of ferroptosis, a new pathway that regulates cell death characterized by the iron-dependent accumulation of lipid hydroperoxides. Interaction between VDAC2/3 and Erastin results in degradation of the channels following activation of ubiquitin protein ligase Nedd4 [38].
Finally, the VDAC3 isoform is associated with cytosolic proteins as tubulins and cytoskeletal proteins, stress sensors, chaperones, and proteasome components, redox-mediating enzymes such as protein disulfide isomerase [39].
2. Proteomics of VDAC Isoforms
2.1. Sample Preparation
Sample preparation has a profound effect on the final results of a proteomic workflow. Protein extraction methods and protein separation techniques should provide an unbiased and reliable map representative of all proteins present in a specific sample. The different extraction and fractionation approaches are based on proteins physicochemical and structural characteristics, such as molecular weight, solubility, hydrophobicity, and isoelectric point. A specific protocol has to be optimized for each particular sample, to maximize protein recovery and minimize the possible proteolysis and amino acid modifications. For these reasons, there is no universal extraction protocol and not a unique buffer composition. Regarding the extraction method, the different strategies available need to be compatible with both the amount of the processed material and the subsequent analytical approach (i.e., separation or MS).
The structural characterization of VDACs presents challenging issues due to their very high hydrophobicity, low solubility, and the impossibility to separate them from other mitochondrial proteins of similar hydrophobicity and to easily isolate each single isoform. In fact, isolation of VDACs has been possible exclusively for plant VDAC isoforms by chromatofocusing, thanks to the absence of phosphorylation sites in their structure [40]. Consequently, it is necessary to analyze them as components of a relatively complex mixture.
A bottom-up proteomic approach was used to investigate the VDAC3 from rat liver mitochondria (rVDAC3) [41]. According with a standard procedure [42], mitochondria were extracted and lysed with a buffer containing 3% Triton X-100 at pH 7.0. The VDAC proteins were partially purified by hydroxyapatite (HTP) chromatography, which allows to obtain a VDACs enriched fraction which comprises also other mitochondria hydrophobic proteins. After precipitation with cold acetone, the protein pellet was solubilized in SDS buffer and loaded on a 17% polyacrylamide gel (1D-SDS-PAGE). The bands in the range 30–35 kDa were manually excised from the gel, cut in small pieces, and subjected to reduction with DTT and alkylation by addition of IAA. Finally, the reduced and carboxyamidomethylated proteins were in gel-digested using trypsin and chymotrypsin, and the resulting peptide mixtures were analyzed by nUHPLC/HRMS [41]. MS data showed that rVDAC3 was found in the whole range 30–35 kDa, together with other proteins, mainly VDAC1, VDAC2, and several other mitochondrial proteins. The reason for VDAC3 electrophoretic heterogeneity probably stems from (i) the different pattern of cysteine oxidations that can modify the protein mobility; (ii) the different amount and quality of cysteine oxidations in various molecules (“redox isomers”).
The gel-digestion procedure shows some disadvantages: (i) larger peptides can get trapped between the gel meshes and lost during the extraction phase of the peptides from the gel; (ii) the electrophoretic procedure itself could damage the samples and alter the redox state of the sulfur amino acids (due to possible over heating generated by the applied voltage and to the presence of residual quantities of the catalysts used for the polyacrylamide polymerization). Furthermore, electrophoresis requires a relatively high amount of sample and the utilization of dyes and detergents. These last molecules could interfere with subsequent MS analyses because these compounds are difficult to eliminate from the sample.
2-DE could potentially represent a useful alternative to 1-DE to improve the separation of VDAC isoforms, but its utilization presents other problems. Actually, this kind of proteins has been under-represented in 2-DE gels due to difficulties in extracting and solubilizing them in the isoelectric focusing sample buffer. In fact, the most effective solubilizing agent for highly hydrophobic membrane proteins is SDS, but this detergent is incompatible with 2-DE. In addition to the difficulties in entering IPG (immobilized pH gradient) gels, membrane proteins tend to precipitate at their isoelectric point during IEF. Furthermore, their tendency to absorb the IPG matrix prevents their migration into the SDS-PAGE gel.
An improvement in the rVDAC3 mass spectrometric analysis was obtained following the introduction of a gel-free shotgun proteomic approach [41]. According to this procedure, to avoid any possible artefact due to air exposure and manipulations, reduction/alkylation was carried out before VDACs purification from the mitochondria. Afterwards, all the proteins present in the HTP eluate, without previous electrophoretic separation, were purified from non-protein contaminating molecules with the PlusOne 2-D Clean-Up kit, and the desalted protein pellet was then re-dissolved in ammonium bicarbonate containing RapiGest SF to improve the solubility. In fact, this surfactant makes the proteins more susceptible to enzymatic cleavage without modifying the sample or inhibiting endoprotease activity. Furthermore, the RapiGest SF is compatible with enzymes such as trypsin or chymotrypsin and does not influence subsequent MS analysis because it can be easily removed in acidic conditions.
Separate aliquots of reduced and alkylated proteins were then subjected to digestion with modified porcine trypsin and chymotrypsin. In this experiment, every protein in the HTP eluate was digested, producing a very complex peptide mixture, which was finally analyzed by nUHPLC/HR nESI-MS/MS.
The new “in solution-digestion” protocol associated with nUHPLC/HR ESI-MS/MS allowed to extend the coverage of the rat and human VDACs sequences with respect to that obtained with the previous procedure [43], so that it was possible to completely cover the rat and human VDAC1 sequences and almost completely the rat and human VDAC2 and VDAC3 sequences [22][41][44]. It should be noted that the short regions not identified in VDAC2 and VDAC3 correspond to small tryptic or chymotryptic peptides or even to single amino acids, which cannot be detected in LC/MS analysis.
Moreover, by means of this new procedure a detailed characterization of PTMs of the three VDACs was obtained (see next paragraphs).
2.2. Mass Spectrometry Analysis of Post-Translational Modifications
The mammalian proteome is vastly more complex than the related genome. The reasons for this difference reside both in the molecular mechanisms that allow a single gene to encode for multiple proteins (genomic recombination, transcription initiation at alternative promoters, differential transcription termination, and alternative splicing of the transcript) and in the post-translational modifications (PTMs) which represent a wide range of chemical changes that proteins can undergo after synthesis. They include the specific cleavage of protein precursors, the covalent addition or removal of low-molecular weight groups (i.e., acetylation, glycosylation, hydroxylation, phosphorylation, ubiquitination) and the formation of disulfide bonds or other redox modifications [45][46][47].
PTMs play crucial roles in cell biology since they can change protein physical or chemical properties, activity, localization, and/or stability. Traditionally, PTMs have been identified by Edman degradation, amino acid analysis, isotopic labeling, or immunochemistry. Within recent years, MS has proven to be extremely useful in PTM discovery. Post-translationally modified amino acids always have a different molecular mass than the original, unmodified residues and this mass increment or deficit is usually the basis for the detection and characterization of PTM by MS (commonly by LC-ESI-MS/MS).
MS has several advantages for characterization of PTMs, including (i) very high sensitivity; (ii) discovery of novel PTMs; (iii) ability to identify PTMs and the modified sites, even in complex protein mixtures; and (iv) ability to quantify the relative changes in PTM occupancy at distinct sites. None of the other techniques provide all these features, so the greater majority of the known PTMs have been described by MS [48].
To improve sensibility, several methods have been developed to enrich the samples in proteins or peptides with specific PTMs prior to MS/MS analysis, such as anti-pY antibodies, IMAC (immobilized metal affinity chromatography) and TiO2 for phosphorylation [49][50], affinity capture with lectins for glycosylated proteins [51], and resin coupled with anti-acetyl-lysine for acetylated proteins [52]. Although, as previously described, isolation of single isoforms of VDACs cannot be obtained, application of combined HPLC and high-resolution ESI-MSMS analysis has resulted in the identification of several PTM in these proteins. In the following, a summary of the MS-based PTMs characterized in VDACs is reported and the respective biological significance discussed. These results are resumed in Table 1 and Figure 1.
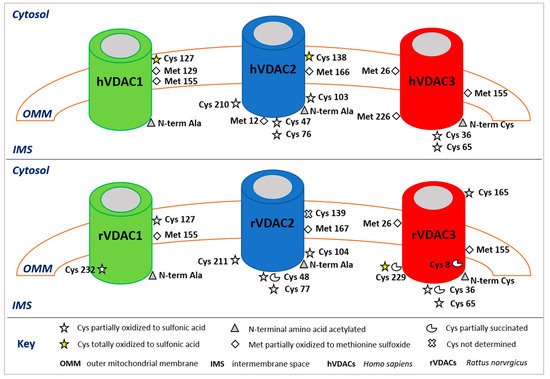
Figure 1. Post-translational modifications of human (upper panel) and rattus (lower panel) VDAC isoforms. The image shows only the modified amino acids and their positions with respect to the cytosol, the outer mitochondrial membrane (OMM), and the intermembrane space (IMS). In the rVDAC1 Cys232 faces the aqueous inside of the pore; in the rVDAC3 Cys8 is located inside of the pore.
Table 1. Post-translational modifications in VDAC isoforms obtained using mass spectrometry. PTM type, mass shift (Da), source of the sample, modified residue, MS method and relative reference are reported. Studies are described by listing first author + year.
| ISOFORM | PTM Type | ΔMass (Da) | Source | Residue | Method | Study |
|---|---|---|---|---|---|---|
| VDAC1 | Protein N-terminal acetylation | 42.0106 | Rat liver | Ala 2 | nUHPLC/high resolution nESI-MS/MS in a Q-QT-qIT MS |
Saletti et al., 2018 |
| HAP1 cells | Ala 2 | Pittalà et al., 2020 | ||||
| Acetylation | 42.0106 | Mouse liver | Lys 33, 41, 74, 234 | nHPLC MS/MS in an LTQ MS | Kim et al., 2006 | |
| Lys 41, 122, 132 | nHPLC MS/MS in an LTQ 2D ion-trap MS |
Schwer et al., 2009 | ||||
| Mouse liver and heart | Lys 237 | UPLC Velos-FT MS | Yang et al., 2011 | |||
| Human liver | Lys 28 | LC/LC-MS/MS in an FTICR/MS | Zhao et al., 2010 | |||
| Oxidation | 15.9949 | Rat liver | Met 155 | LC/LC-MS/MS in an FTICR/MS | Guan et al., 2003 | |
| nUHPLC/high resolution nESI-MS/MS in a Q-QT-qIT MS |
Saletti et al., 2018 | |||||
| HAP1 cells | Met 129, 155 | Pittalà et al., 2020 | ||||
| Trioxidation | 47.9847 | Rat liver | Cys 127, 232 | Saletti et al., 2018 | ||
| HAP1 cells | Cys 127 | Pittalà et al., 2020 | ||||
| Phosphorylation | 79.9663 | Rat liver | Ser 12, 136 | HPLC MS/MS in an LTQ MS | Distler et al., 2007 | |
| Mouse liver | Ser 117 | nHPLC MS/MS in an LTQ MS | Lee et al., 2007 | |||
| HeLa cells | Ser 101, 102, 104, Thr 107 |
nHPLC MS/MS in an LTQ-Orbitrap MS |
Olsen et al., 2006 | |||
| Mouse brain | Tyr 80, 208 | LC-MS/MS in an LTQ FT MS | Ballif et al., 2008 | |||
| VDAC2 | Protein N-terminal acetylation | 42.0106 | Rat liver | Ala 2 | nUHPLC/high resolution nESI-MS/MS in a Q-QT-qIT MS |
Saletti et al., 2018 |
| HAP1 cells | Ala 2 | Pittalà et al., 2020 | ||||
| Acetylation | 42.0106 | Mouse liver | Lys 32, 75 | nHPLC MS/MS in an LTQ MS | Kim et al., 2006 | |
| Lys 121 | nHPLC MS/MS in an LTQ 2D ion-trap MS |
Schwer et al., 2009 | ||||
| Oxidation | 15.9949 | Rat liver | Met 167 | nUHPLC/high resolution nESI-MS/MS in a Q-QT-qIT MS |
Saletti et al., 2018 | |
| HAP1 cells | Met 12, 166 | Pittalà et al., 2020 | ||||
| Trioxidation | 47.9847 | Rat liver | Cys 48, 77, 104, 211 | Saletti et al., 2018 | ||
| HAP1 cells | Cys 47, 76, 103, 138, 210 | Pittalà et al., 2020 | ||||
| Succination | 116.0110 | Mouse brain | Cys 48, 77 | LC-nESI-MS/MS in an LTQ-Orbitrap MS |
Piroli et al., 2016 | |
| Rat liver | Cys 48 | nUHPLC/high resolution nESI-MS/MS in a Q-QT-qIT MS |
Saletti et al., 2018 | |||
| Phosphorylation | 79.9663 | HeLa cells | Ser 115, Thr 118 | nHPLC MS/MS in an LTQ-Orbitrap MS |
Olsen et al., 2006 | |
| Rat liver | Thr 109 | SCX-RP-MS/MS in an LTQ-Orbitrap MS |
Deng et al., 2010 | |||
| Rat liver | Tyr 237 | HPLC MS/MS in an LTQ MS | Distler et al., 2007 | |||
| Mouse brain | Tyr 207 | LC-MS/MS in an LTQ FT MS | Ballif et al., 2008 | |||
| VDAC3 | Protein N-terminal acetylation | 42.0106 | Rat liver | Cys 2 | nUHPLC/high resolution nESI-MS/MS in a Q-QT-qIT MS |
Saletti et al., 2016 |
| HAP1 cells | Cys 2 | Pittalà et al., 2020 | ||||
| Acetylation | 42.0106 | Mouse liver | Lys 20, 61, 226 | nHPLC MS/MS in an LTQ MS | Kim et al., 2006 | |
| Lys 63, 109 | nHPLC MS/MS in an LTQ 2D ion-trap MS |
Schwer et al., 2009 | ||||
| Human liver | Lys 28 | LC/LC-MS/MS in an FTICR-MS | Zhao et al., 2010 | |||
| Oxidation | 15.9949 | Rat liver | Met 26, 155 | nUHPLC/high resolution nESI-MS/MS in a Q-QT-qIT MS |
Saletti et al., 2016 | |
| HAP1 cells | Met 26, 155, 226 | Pittalà et al., 2020 | ||||
| Trioxidation | 47.9847 | Rat liver | Cys 36, 65, 165, 229 | Saletti et al., 2016 | ||
| HAP1 cells | Cys 36, 65 | Pittalà et al., 2020 | ||||
| Succination | 116.0110 | Rat liver | Cys 8, 36, 229 | Saletti et al., 2018 | ||
| Phosphorylation | 79.9663 | Rat liver | Ser 241, Thr 33 | HPLC MS/MS in an LTQ MS | Distler et al., 2007 | |
| Mouse brain | Tyr 49 | LC-MS/MS in an LTQ FT MS | Ballif et al., 2008 |
This entry is adapted from the peer-reviewed paper 10.3390/ijms222312833
References
- Shoshan-Barmatz, V.; De Pinto, V.; Zweckstetter, M.; Raviv, Z.; Keinan, N.; Arbel, N. VDAC, a multi-functional mitochon-drial protein regulating cell life and death. Mol. Aspects Med. 2010, 31, 227–285.
- Messina, A.; Reina, S.; Guarino, F.; De Pinto, V. VDAC isoforms in mammals. Biochim. Biophys. Acta 2012, 1818, 1466–1476.
- Young, M.J.; Bay, D.C.; Hausner, G.; Court, D.A. The evolutionary history of mitochondrial porins. BMC Evol. Biol. 2007, 7, 31.
- De Pinto, V.; Reina, S.; Gupta, A.; Messina, A.; Mahalakshmi, R. Role of cysteines in mammalian VDAC isoforms’ function. Biochim. Biophys. Acta Bioenerg. 2016, 1857, 1219–1227.
- Hiller, S.; Garces, R.G.; Malia, T.J.; Orekhov, V.Y.; Colombini, M.; Wagner, G. Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science 2008, 321, 1206–1210.
- Bayrhuber, M.; Meins, T.; Habeck, M.; Becker, S.; Giller, K.; Villinger, S.; Vonrhein, C.; Griesinger, C.; Zweckstetter, M.; Zeth, K. Structure of the human voltage-dependent anion channel. Proc. Natl. Acad. Sci. USA 2008, 105, 15370–15375.
- Ujwal, R.; Cascio, D.; Colletier, J.P.; Faham, S.; Zhang, J.; Toro, L.; Ping, P.; Abramson, J. The crystal structure of mouse VDAC1 at 2.3 angstrom resolution reveals mechanistic insights into metabolite gating. Proc. Natl. Acad. Sci. USA 2008, 105, 17742–17747.
- Schredelseker, J.; Paz, A.; Lopez, C.J.; Altenbach, C.; Leung, C.S.; Drexler, M.K.; Chen, J.N.; Hubbell, W.L.; Abramson, J. High resolution structure and double electron-electron resonance of the zebrafish voltage-dependent anion channel 2 reveal an oligomeric population. J. Biol. Chem. 2014, 289, 12566–12577.
- Amodeo, G.F.; Scorciapino, M.A.; Messina, A.; De Pinto, V.; Ceccarelli, M. Charged residues distribution modulates selec-tivity on the open state of human isoforms of the voltage dependent anion-selective channel. PLoS ONE 2014, 9, e103879.
- De Pinto, V.; Guarino, F.; Guarnera, A.; Messina, A.; Reina, S.; Tomasello, M.F.; Palermo, V.; Mazzoni, C. Characterization of human VDAC isoforms: A peculiar function for VDAC3? Biochim. Biophys. Acta-Bioenergetics 2010, 1797, 1268-1275.
- Naghdi, S.; Várnai, P.; Hajnóczky, G. Motifs of VDAC2 required for mitochondrial Bak import and tBid-induced apoptosis. Proc. Natl. Acad. Sci. USA 2015, 112, E5590–E5599.
- Shoshan-Barmatz, V.; Maldonado, E.N.; Krelin, Y. VDAC1 at the crossroads of cell metabolism, apoptosis and cell stress. Cell stress 2017, 1, 1.
- Shoshan-Barmatz, V.; Pittala, S.; Mizrachi, D. VDAC1 and the TSPO: Expression, Interactions, and Associated Functions in Health and Disease States. Int. J. Mol. Sci. 2019, 20, 3348.
- Kerner, J.; Lee, K.; Tandler, B.; Hoppel, C.L. VDAC proteomics: Post-translation modifications. Biochim. Biophys. Acta 2012, 1818, 1520–1525.
- Geula, S.; Ben-Hail, D.; Shoshan-Barmatz, V. Structure-based analysis of vdac1: N-terminus location, translocation, channel gating and association with anti-apoptotic proteins. Biochem. J. 2012, 444, 475–485.
- Abu-Hamad, S.; Arbel, N.; Calo, D.; Arzoine, L.; Israelson, A.; Keinan, N.; Ben-Romano, R.; Friedman, O.; Shoshan-Barmatz, V. The vdac1 n-terminus is essential both for apoptosis and the protective effect of anti-apoptotic proteins. J. Cell Sci. 2009, 122, 1906–1916.
- Shi, Y.; Chen, J.;Weng, C.; Chen, R.; Zheng, Y.; Chen, Q.; Tang, H. Identification of the protein-protein contact site and inter-action mode of human vdac1 with bcl-2 family proteins. Biochem. Biophys. Res. Commun. 2003, 305, 989–996.
- Naghdi, S.; Hajnóczky, G. VDAC2-specific cellular functions and the underlying structure. Biochim. Biophys. Acta 2016, 1863, 2503–2514.
- Shoshan-Barmatz, V.; Ben-Hail, D. VDAC, a multi-functional mitochondrial protein as a pharmacological target. Mitochon-drion 2012, 12, 24–34.
- Abu-Hamad, S.; Zaid, H.; Israelson, A.; Nahon, E.; Shoshan-Barmatz, V. Hexokinase-I protection against apoptotic cell death is mediated via interaction with the voltage-dependent anion channel-1: Mapping the site of binding. J. Biol. Chem. 2008, 283, 13482–13490.
- De Pinto, V.; al Jamal, J.A.; Palmieri, F. Location of the dicyclohexylcarbodiimide-reactive glutamate residue in the bovine heart mitochondrial porin. J. Biol. Chem. 1993, 268, 12977–12982.
- Pittalà, M.G.G.; Saletti, R.; Reina, S.; Cunsolo, V.; De Pinto, V.; Foti, S. A High Resolution Mass Spectrometry Study Reveals the Potential of Disulfide Formation in Human Mitochondrial Voltage-Dependent Anion Selective Channel Isoforms (hVDACs). Int. J. Mol. Sci. 2020, 21, 1468.
- Budelier, M.M.; Cheng, W.W.L.; Bergdoll, L.; Chen, Z.W.; Janetka, J.W.; Abramson, J.; Krishnan, K.; Mydock-McGrane, L.; Covey, D.F.; Whitelegge, J.P.; et al. Photoaffinity labeling with cholesterol analogues precisely maps a cholesterol-binding site in voltage-dependent anion channel-1. J. Biol. Chem. 2017, 292, 9294–9304.
- Gatliff, J.; East, D.; Crosby, J.; Abeti, R.; Harvey, R.; Craigen, W.; Parker, P.; Campanella, M. Tspo interacts with vdac1 and triggers a ros-mediated inhibition of mitochondrial quality control. Autophagy 2015, 10, 2279–2296.
- Mueller, B.K.; Subramaniam, S.; Senes, A. A frequent, GxxxG-mediated, transmembrane association motif is optimized for the formation of interhelical Cα-H hydrogen bonds. Proc. Natl. Acad. Sci. USA 2014, 111, E888–E895.
- Bernardi, P.; Di Lisa, F.; Fogolari, F.; Lippe, G. From ATP to PTP and back: A dual function for the mitochondrial ATP syn-thase. Circ. Res. 2015, 116, 1850–1862.
- Reina, S.; Guarino, F.; Magrì, A.; De Pinto, V. VDAC3 as a potential marker of mitochondrial status is involved in cancer and pathology. Front. Oncol. 2016, 6, 264.
- Magrì, A.; Messina, A. Interactions of VDAC with Proteins Involved in Neurodegenerative Aggregation: An Opportunity for Advancement on Therapeutic Molecules. Curr. Med. Chem. 2017, 24, 4470–4487.
- Sun, Y.; Vashisht, A.A.; Tchieu, J.; Wohlschlegel, J.A.; Dreier, L. Voltage-dependent Anion Channels (VDACs) recruit Parkin to defective mitochondria to promote mitochondrial autophagy. J. Biol. Chem. 2012, 287, 40652–40660.
- Sheikh, S.; Safia; Haque, E.; Snober, S.M. Neurodegenerative diseases: Multifactorial conformational diseases and their therapeutic interventions. J. Neurodegen. Dis. 2013, 2013, 563481.
- Smilansky, A.; Dangoor, L.; Nakdimon, I.; Ben-Hail, D.; Mizrachi, D.; Shoshan-Barmatz, V. The voltage-dependent anion channel 1 mediates amyloid beta toxicity and represents a potential target for Alzheimer's disease therapy. J. Biol. Chem. 2015, 290, 30670–30683.
- Hemachandra Reddy, P. Is the mitochondrial outer membrane protein VDAC1 therapeutic target for Alzheimer’s disease? Biochim. Biophys. Acta 2013, 1832, 67–75.
- Rostovtseva, T.K.; Gurnev, P.A.; Protchenko, O.; Hoogerheide, D.P.; Yap, T.L.; Philpott, C.C.; Lee, J.C.; Bezrukov, S.M. α-Synuclein shows high affinity interaction with Voltage-dependent Anion Channel, suggesting mechanisms of mitochon-drial regulation and toxicity in Parkinson Disease. J. Biol. Chem. 2015, 290, 18467–18477.
- Israelson, A.; Arbel, N.; Da Cruz, S.; Ilieva, H.; Yamanaka, K.; Shoshan-Barmatz, V.; Cleveland, D.W. Misfolded mutant SOD1 directly inhibits VDAC1 conductance in a mouse model of inherited ALS. Neuron 2010, 67, 575–587.
- Magrì, A.; Belfiore, R.; Reina, S.; Tomasello, M.F.; Di Rosa, M.C.; Guarino, F.; Leggio, L.; De Pinto, V.; Messina, A. Hexoki-nase I N-terminal based peptide prevents the VDAC1-SOD1G93A interaction and re-establishes ALS cell viability. Sci. Rep. 2016, 6, 34802.
- Harada, T.; Sada, R.; Osugi, Y.; Matsumoto, S.; Matsuda, T.; Hayashi-Nishino, M.; Nagai, T.; Harada, A.; Kikuchi, A. Pal-mitoylated CKAP4 regulates mitochondrial functions through an interaction with VDAC2 at ER–mitochondria contact sites. J. Cell Sci. 2020, 133, jcs249045.
- Zhong, Y.; Tang, X.; Sheng, X.; Xing, J.; Zhan, W. Voltage-Dependent Anion Channel Protein 2 (VDAC2) and Receptor of Ac-tivated Protein C Kinase 1 (RACK1) Act as Functional Receptors for Lymphocystis Disease Virus Infection. J. Virol. 2019, 93, e00122-19.
- Yang, Y.; Luo, M.; Zhang, K.; Zhang, J.; Gao, T.; O’Connell, D.; Yao, F.; Mu, C.; Cai, B.; Shang, Y.; Chen, W. Nedd4 ubiq-uitylates VDAC2/3 to suppress erastin-induced ferroptosis in melanoma. Nat. Commun. 2020, 11, 433.
- Messina, A.; Reina, S.; Guarino, F.; Magrì, A.; Tomasello, F.; Clark, R.E.; Ramsayc, R.R.; De Pinto, V. Live cell interactome of the human voltage dependent anion channel 3 (VDAC3) revealed in HeLa cells by affinity purification tag technique. Mol. BioSyst. 2014, 10, 2134–2145.
- Abrecht, H.; Wattiez, R.; Ruysschaert, J.M.; Homblé, F. Purification and characterization of two Voltage-Dependent Anion Channel Isoforms from plant seeds. Plant Physiol. 2000, 124, 1181–1190.
- Saletti, R.; Reina, S.; Pittalà, M.G.G.; Belfiore, R.; Cunsolo, V.; Messina, A.; De Pinto, V.; Foti, S. High resolution mass spec-trometry characterization of the oxidation pattern of methionine and cysteine residues in rat liver mitochondria Volt-age-Dependent Anion selective Channel 3 (VDAC3). Biochim. Biophys. Acta-Biomembr. 2017, 1859, 301–311.
- De Pinto, V.; Prezioso, G.; Palmieri, F. A simple and rapid method for the purification of the mitochondrial porin from mammalian tissues. Biochim. Biophys. Acta 1987, 905, 499–502.
- Distler, A.M.; Kerner, J.; Peterman, S.M.; Hoppel, C.L. A targeted proteomic approach for the analysis of rat liver mito-chondrial outer membrane proteins with extensive sequence coverage. Anal. Biochem. 2006, 356, 18–29.
- Saletti, R.; Reina, S.; Pittalà, M.G.G.; Magrì, A.; Cunsolo, V.; Foti, S.; De Pinto, V. Post-translational modifications of VDAC1 and VDAC2 cysteines from rat liver mitochondria. Biochim. Biophys. Acta-Bioenerg. 2018, 1859, 806–816.
- Wang, Y.; Peterson, S.; Loring, J. Protein post-translational modifications and regulation of pluripotency in human stem cells. Cell Res. 2014, 24, 143–160.
- Duan, G.; Walther, D. The roles of post-translational modifications in the context of protein interaction networks. PLoS Comput. Biol. 2015, 11, e1004049.
- Reina, S.; Pittalà, M.G.G.; Guarino, F.; Messina, A.; De Pinto, V.; Foti, S.; Saletti, R. Cysteine oxidations in mitochondrial membrane proteins: The case of VDAC isoforms in mammals. Front. Cell Dev. Biol. 2020, 8, 397.
- Jensen, O.N. Modification-specific proteomics: Characterization of posttranslational modifications by mass spectrometry. Curr. Opin. Chem. Biol. 2004, 8, 33–41.
- Corthals, G.L.; Aebersold, R.; Goodlett, D.R. Identification of phosphorylation sites using microimmobilized metal affinity chromatography. Meth. Enzymol. 2005, 405, 66–81.
- Larsen, M.R.; Thingholm, T.E.; Jensen, O.N.; Roepstorff, P.; Jørgensen, T.J. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol. Cell. Proteomics 2005, 4, 873–886.
- Yang, Z.; Hancock, W.S. Approach to the comprehensive analysis of glycoproteins isolated from human serum using a mul-ti-lectin affinity column. J. Chromatogr. A 2004, 1053, 79–88.
- Kim, S.C.; Sprung, R.; Chen, Y.; Xu, Y.; Ball, H.; Pei, J.; Cheng, T.; Kho, Y.; Xiao, H.; Xiao, L.; et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell. 2006, 23, 607–618.
This entry is offline, you can click here to edit this entry!
