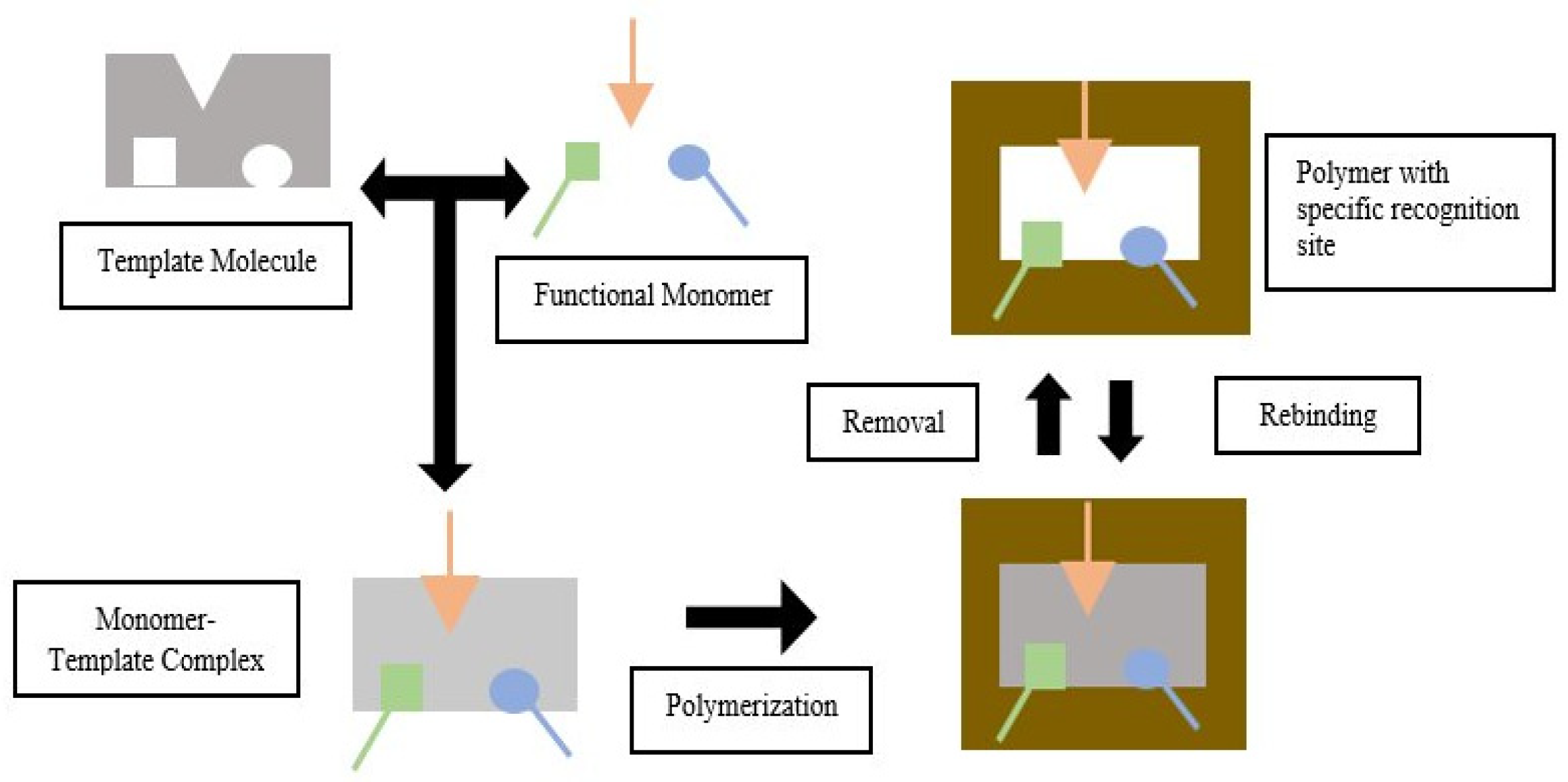
Molecular imprinting is a technique for creating artificial recognition sites on polymer matrices that complement the template in terms of size, shape, and spatial arrangement of functional groups. The main advantage of Molecularly Imprinted Polymers (MIP) as the polymer for use with a molecular imprinting technique is that they have high selectivity and affinity for the target molecules used in the molding process.
- molecular imprinted polymer
- interaction mechanism
- template-monomer interaction
- MIP-template interaction
1. Introduction
2. MIP Application
2.1. Environmental Monitoring
2.2. Food Analysis
2.3. Biomedical Diagnostic
2.4. Drug Delivery
3. Choosing Right Component for MIP
3.1. Functional Monomers
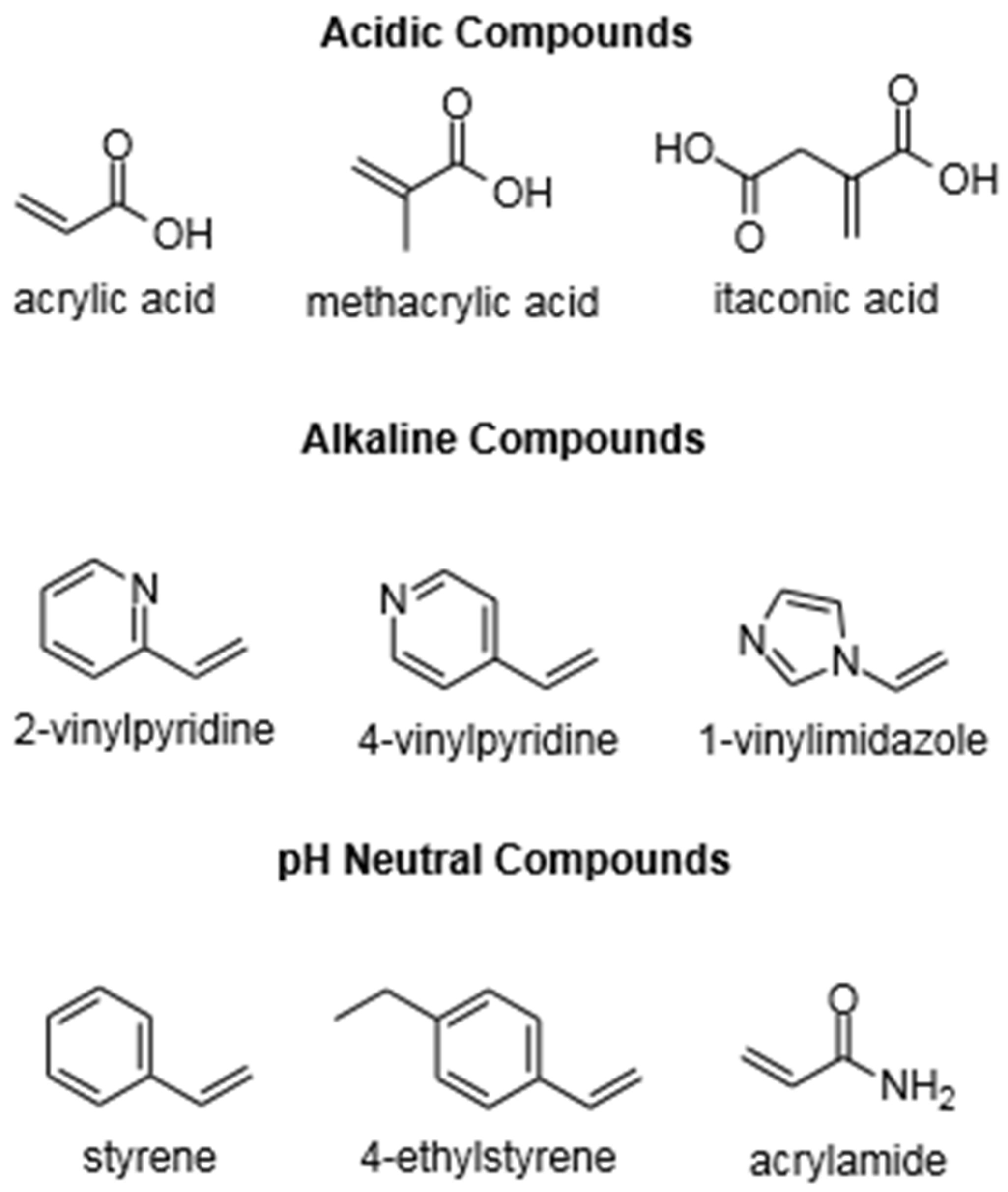
3.2. Cross-Linker
3.3. Solvents
4. Template-Monomer Interaction
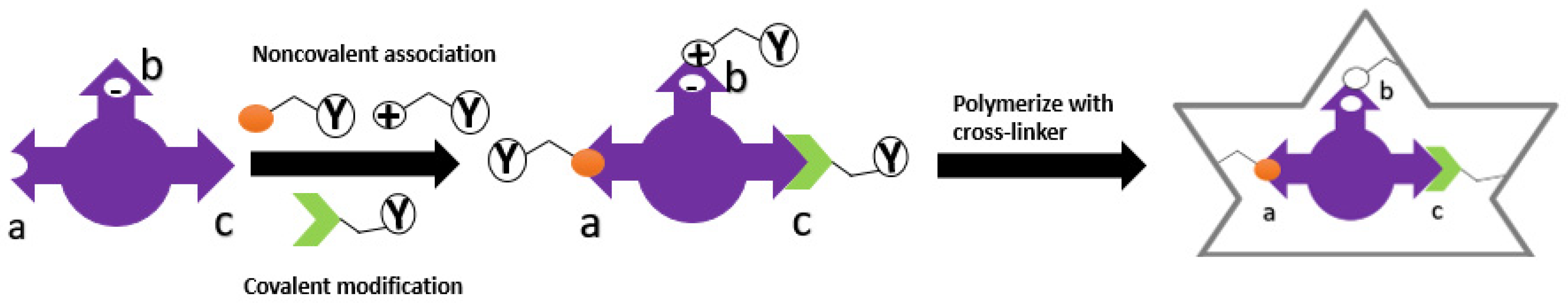
|
Imprinting Type |
Covalent |
Non-Covalent |
References |
|---|---|---|---|
|
Interaction |
|
|
|
|
Advantage |
|
|
|
|
Disadvantage |
|
|
5. Analysis of Template-Monomer Functional Interactions
This entry is adapted from the peer-reviewed paper 10.3390/molecules26185612
References
- Ertürk, G.; Mattiasson, B. Molecular imprinting techniques used for the preparation of biosensors. Sensors 2017, 17, 288.
- Sanbe, H.; Haginaka, J. Uniformly sized molecularly imprinted polymers for bisphenol A and β-estradiol: Retention and molecular recognition properties in hydro-organic mobile phases. J. Pharm. Biomed. Anal. 2003, 30, 1835–1844.
- Abo Dena, A.S.; Ali, A.M.; El-Sherbiny, I. Surface-Imprinted Polymers (SIPs): Advanced Materials for Bio-Recognition. J. Nanotechnol. Adv. Mater. 2020, 8, 1–19.
- Kalecki, J.; Iskierko, Z.; Cieplak, M.; Sharma, P.S. Oriented Immobilization of Protein Templates: A New Trend in Surface Imprinting. ACS Sens. 2020, 5, 3710–3720.
- Yongabi, D.; Khorshid, M.; Losada-Pérez, P.; Eersels, K.; Deschaume, O.; D’Haen, J.; Bartic, C.; Hooyberghs, J.; Thoelen, R.; Wübbenhorst, M.; et al. Cell detection by surface imprinted polymers SIPs: A study to unravel the recognition mechanisms. Sens. Actuators B Chem. 2018, 255, 907–917.
- Idil, N.; Bakhshpour, M.; Perçin, I.; Mattiasson, B. Whole Cell Recognition of Staphylococcus aureus Using Biomimetic SPR Sensors. Biosensors 2021, 11, 140.
- Pasquardini, L.; Bossi, A.M. Molecularly imprinted polymers by epitope imprinting: A journey from molecular interactions to the available bioinformatics resources to scout for epitope templates. Anal. Bioanal. Chem. 2021, 413, 6101–6115.
- Trends, T.; Chemistry, A.; Merko, A. New materials for electrochemical sensing IV. Molecular imprinted polymers. TrAC Trends Anal. Chem. 2016, 21, 717–725.
- Sellergren, B.; Allender, C.J. Molecularly imprinted polymers: A bridge to advanced drug delivery. Adv. Drug Deliv. Rev. 2005, 57, 1733–1741.
- Chen, L.; Wang, X.; Lu, W.; Wu, X.; Li, J. Molecular imprinting: Perspectives and applications. Chem. Soc. Rev. 2016, 45, 2137–2211.
- Vasapollo, G.; Del Sole, R.; Mergola, L.; Lazzoi, M.R.; Scardino, A.; Scorrano, S.; Mele, G. Molecularly imprinted polymers: Present and future prospective. Int. J. Mol. Sci. 2011, 12, 5908–5945.
- Guć, M.; Schroeder, G. Application of Molecularly Imprinted Polymers (MIP) and Magnetic Molecularly Imprinted Polymers (mag-MIP) to Selective Analysis of Quercetin in Flowing Atmospheric-Pressure Afterglow Mass Spectrometry (FAPA-MS) and in Electrospray Ionization Mass Spectrom. Molecules 2019, 24, 2364.
- Arifuzzaman, M.; Zhao, Y. Water-Soluble Molecularly Imprinted Nanoparticle Receptors with Hydrogen-Bond-Assisted Hydrophobic Binding. J. Org. Chem. 2016, 81, 7518–7526.
- Rebelo, P.; Costa-Rama, E.; Seguro, I.; Pacheco, J.G.; Nouws, H.P.A.; Cordeiro, M.N.D.S.; Delerue-Matos, C. Molecularly imprinted polymer-based electrochemical sensors for environmental analysis. Biosens. Bioelectron. 2021, 172, 112719.
- Okutucu, B. Wastewater Treatment Using Imprinted Polymeric Adsorbents. In Waste in Textile and Leather Sectors; IntechOpen: London, UK, 2020.
- Ren, X.; Zeng, G.; Tang, L.; Wang, J.; Wan, J.; Liu, Y.; Yu, J.; Yi, H.; Ye, S.; Deng, R. Sorption, transport and biodegradation—An insight into bioavailability of persistent organic pollutants in soil. Sci. Total Environ. 2018, 610–611, 1154–1163.
- Farooq, S.; Nie, J.; Cheng, Y.; Yan, Z.; Li, J.; Bacha, S.A.S.; Mushtaq, A.; Zhang, H. Molecularly imprinted polymers’ application in pesticide residue detection. Analyst 2018, 143, 3971–3989.
- Capcarova, M.; Zbynovska, K.; Kalafova, A.; Bulla, J.; Bielik, P. Environment contamination by mycotoxins and their occurrence in food and feed: Physiological aspects and economical approach. J. Environ. Sci. Health Part B Pestic. Food Contam. Agric. Wastes 2016, 51, 236–244.
- Naseri, M.; Mohammadniaei, M.; Sun, Y.; Ashley, J. The use of aptamers and molecularly imprinted polymers in biosensors for environmental monitoring: A tale of two receptors. Chemosensors 2020, 8, 32.
- Yang, Q.; Li, J.; Wang, X.; Peng, H.; Xiong, H.; Chen, L. Strategies of molecular imprinting-based fluorescence sensors for chemical and biological analysis. Biosens. Bioelectron. 2018, 112, 54–71.
- Azizi, A.; Bottaro, C.S. A critical review of molecularly imprinted polymers for the analysis of organic pollutants in environmental water samples. J. Chromatogr. A 2020, 1614, 460603.
- Ndunda, E.N.; Mizaikoff, B. Molecularly imprinted polymers for the analysis and removal of polychlorinated aromatic compounds in the environment: A review. Analyst 2016, 141, 3141–3156.
- Gao, M.; Gao, Y.; Chen, G.; Huang, X.; Xu, X.; Lv, J.; Wang, J.; Xu, D.; Liu, G. Recent Advances and Future Trends in the Detection of Contaminants by Molecularly Imprinted Polymers in Food Samples. Front. Chem. 2020, 8, 1142.
- Song, X.; Zhou, T.; Li, J.; Zhang, M.; Xie, J.; He, L. Determination of ten macrolide drugs in environmental water using molecularly imprinted solid-phase extraction coupled with liquid chromatography-tandem mass spectrometry. Molecules 2018, 23, 1172.
- Garcia, R.; Cabrita, M.J.; Costa Freitas, A.M. Application of Molecularly Imprinted Polymers for the Analysis of Pesticide Residues in Food—A Highly Selective and Innovative Approach. Am. J. Anal. Chem. 2011, 2, 16–25.
- Song, X.; Xu, S.; Chen, L.; Wei, Y.; Xiong, H. Recent advances in molecularly imprinted polymers in food analysis. J. Appl. Polym. Sci. 2014, 131, 40766.
- Garcia, R.; Carreiro, E.P.; Lima, J.C.; da Silva, M.G.; Freitas, A.M.C.; Cabrita, M.J. Assessment of dimethoate in olive oil samples using a dual responsive molecularly imprinting-based approach. Foods 2020, 9, 618.
- Liu, Y.; Yang, Q.; Chen, X.; Song, Y.; Wu, Q.; Yang, Y.; He, L. Sensitive analysis of trace macrolide antibiotics in complex food samples by ambient mass spectrometry with molecularly imprinted polymer-coated wooden tips. Talanta 2019, 204, 238–247.
- Cheubong, C.; Yoshida, A.; Mizukawa, Y.; Hayakawa, N.; Takai, M.; Morishita, T.; Kitayama, Y.; Sunayama, H.; Takeuchi, T. Molecularly Imprinted Nanogels Capable of Porcine Serum Albumin Detection in Raw Meat Extract for Halal Food Control. Anal. Chem. 2020, 92, 6401–6407.
- Negarian, M.; Mohammadinejad, A.; Mohajeri, S.A. Preparation, evaluation and application of core–shell molecularly imprinted particles as the sorbent in solid-phase extraction and analysis of lincomycin residue in pasteurized milk. Food Chem. 2019, 288, 29–38.
- Zhao, Q.; Li, H.; Xu, Y.; Zhang, F.; Zhao, J.; Wang, L.; Hou, J.; Ding, H.; Li, Y.; Jin, H.; et al. Determination triazine pesticides in cereal samples based on single-hole hollow molecularly imprinted microspheres. J. Chromatogr. A 2015, 1376, 26–34.
- Sun, X.; Wang, J.; Li, Y.; Yang, J.; Jin, J.; Shah, S.M.; Chen, J. Novel dummy molecularly imprinted polymers for matrix solid-phase dispersion extraction of eight fluoroquinolones from fish samples. J. Chromatogr. A 2014, 1359, 1–7.
- Pan, J.; Chen, W.; Ma, Y.; Pan, G. Molecularly imprinted polymers as receptor mimics for selective cell recognition. Chem. Soc. Rev. 2018, 47, 5574–5587.
- Zhang, H. Molecularly Imprinted Nanoparticles for Biomedical Applications. Adv. Mater. 2019, 32, 1806328.
- El-Schich, Z.; Zhang, Y.; Feith, M.; Beyer, S.; Sternbæk, L.; Ohlsson, L.; Stollenwerk, M.; Wingren, A.G. Molecularly imprinted polymers in biological applications. Biotechniques 2020, 69, 407–420.
- Takeuchi, T.; Sunayama, H. Beyond natural antibodies—A new generation of synthetic antibodies created by post-imprinting modification of molecularly imprinted polymers. Chem. Commun. 2018, 54, 6243–6251.
- Wang, H.Y.; Cao, P.P.; He, Z.Y.; He, X.W.; Li, W.Y.; Li, Y.H.; Zhang, Y.K. Targeted imaging and targeted therapy of breast cancer cells: Via fluorescent double template-imprinted polymer coated silicon nanoparticles by an epitope approach. Nanoscale 2019, 11, 17018–17030.
- Regan, B.; Boyle, F.; O’Kennedy, R.; Collins, D. Evaluation of molecularly imprinted polymers for point-of-care testing for cardiovascular disease. Sensors 2019, 19, 3485.
- Arshad, R.; Rhouati, A.; Hayat, A.; Nawaz, M.H.; Yameen, M.A.; Mujahid, A.; Latif, U. MIP-Based Impedimetric Sensor for Detecting Dengue Fever Biomarker. Appl. Biochem. Biotechnol. 2020, 191, 1384–1394.
- Selvolini, G.; Marrazza, G. MIP-based sensors: Promising new tools for cancer biomarker determination. Sensors 2017, 17, 718.
- Mayeux, R. Biomarkers: Potential Uses and Limitations. NeuroRx 2004, 1, 182–188.
- Zaidi, S.A. Molecular imprinting: A useful approach for drug delivery. Mater. Sci. Energy Technol. 2020, 3, 72–77.
- Li, C.; Wang, J.; Wang, Y.; Gao, H.; Wei, G.; Huang, Y.; Yu, H.; Gan, Y.; Wang, Y.; Mei, L.; et al. Recent progress in drug delivery. Acta Pharm. Sin. B 2019, 9, 1145–1162.
- Suksuwan, A.; Lomlim, L.; Rungrotmongkol, T.; Nakpheng, T.; Dickert, F.L.; Suedee, R. The composite nanomaterials containing (R)-thalidomide-molecularly imprinted polymers as a recognition system for enantioselective-controlled release and targeted drug delivery. J. Appl. Polym. Sci. 2015, 132, 41930.
- Włoch, M.; Datta, J. Synthesis and polymerisation techniques of molecularly imprinted polymers. In Comprehensive Analytical Chemistry; Elsevier: Amsterdam, The Netherlands, 2019; Volume 86, pp. 17–40.
- Urraca, J.L.; Carbajo, M.C.; Torralvo, M.J.; González-Vázquez, J.; Orellana, G.; Moreno-Bondi, M.C. Effect of the template and functional monomer on the textural properties of molecularly imprinted polymers. Biosens. Bioelectron. 2008, 24, 155–161.
- Barros, L.A.; Custodio, R.; Rath, S. Design of a New Molecularly Imprinted Polymer Selective for Hydrochlorothiazide Based on Theoretical Predictions Using Gibbs Free Energy. J. Braz. Chem. Soc. 2016, 27, 2300–2311.
- Joke Chow, A.L.; Bhawani, S.A. Synthesis and Characterization of Molecular Imprinting Polymer Microspheres of Cinnamic Acid: Extraction of Cinnamic Acid from Spiked Blood Plasma. Int. J. Polym. Sci. 2016, 2016, 2418915.
- Fu, X.; Yang, Q.; Zhou, Q.; Lin, Q.; Wang, C. Template-Monomer Interaction in Molecular Imprinting: Is the Strongest the Best? Open J. Org. Polym. Mater. 2015, 5, 58–68.
- Singh, M.; Singh, S.; Singh, S.P.; Patel, S.S. Recent advancement of carbon nanomaterials engrained molecular imprinted polymer for environmental matrix. Trends Environ. Anal. Chem. 2020, 27, e00092.
- Yu, H.; He, Y.; She, Y.; Wang, M.; Yan, Z.; Ren, J.H.; Cao, Z.; Shao, Y.; Wang, S.; Abd El-Aty, A.M.; et al. Preparation of molecularly imprinted polymers coupled with high-performance liquid chromatography for the selective extraction of salidroside from Rhodiola crenulata. J. Chromatogr. B 2019, 1118–1119, 180–186.
- Sánchez-González, J.; Peña-Gallego, Á.; Sanmartín, J.; Bermejo, A.M.; Bermejo-Barrera, P.; Moreda-Piñeiro, A. NMR spectroscopy for assessing cocaine-functional monomer interactions when preparing molecularly imprinted polymers. Microchem. J. 2019, 147, 813–817.
- Wu, H.; Tian, Q.; Zheng, W.; Jiang, Y.; Xu, J.; Li, X.; Zhang, W.; Qiu, F. Non-enzymatic glucose sensor based on molecularly imprinted polymer: A theoretical, strategy fabrication and application. J. Solid State Electrochem. 2019, 23, 1379–1388.
- Zhong, M.; Wang, Y.-H.; Wang, L.; Long, R.-Q.; Chen, C.-L. Preparation and application of magnetic molecularly imprinted polymers for the isolation of chelerythrine from Macleaya cordata. J. Sep. Sci. 2018, 41, 3318–3327.
- Nikoleli, G.P.; Nikolelis, D.P.; Siontorou, C.G.; Karapetis, S.; Varzakas, T. Novel Biosensors for the Rapid Detection of Toxicants in Foods. Adv. Food Nutr. Res. 2018, 84, 57–102.
- Marć, M.; Wieczorek, P.P. Chapter One—Introduction to MIP synthesis, characteristics and analytical application. In Comprehensive Analytical Chemistry; Elsevier: Amsterdam, The Netherlands, 2019; Volume 86, pp. 1–15.
- Anene, A.; Kalfat, R.; Chevalier, Y.; Hbaieb, S. Design of Molecularly Imprinted Polymeric Materials: The Crucial Choice of Functional Monomers. Chem. Afr. 2020, 3, 769–781.
- Zhao, G.; Liu, J.; Liu, M.; Han, X.; Peng, Y.; Tian, X.; Liu, J.; Zhang, S. Synthesis of molecularly imprinted polymer via emulsion polymerization for application in solanesol separation. Appl. Sci. 2020, 10, 2868.
- Ibarra, I.S.; Miranda, J.M.; Pérez-Silva, I.; Jardinez, C.; Islas, G. Sample treatment based on molecularly imprinted polymers for the analysis of veterinary drugs in food samples: A review. Anal. Methods 2020, 12, 2958–2977.
- Xu, X.; Duhoranimana, E.; Zhang, X. Preparation and characterization of magnetic molecularly imprinted polymers for the extraction of hexamethylenetetramine in milk samples. Talanta 2017, 163, 31–38.
- Zunngu, S.S.; Madikizela, L.M.; Chimuka, L.; Mdluli, P.S. Synthesis and application of a molecularly imprinted polymer in the solid-phase extraction of ketoprofen from wastewater. Comptes Rendus Chim. 2017, 20, 585–591.
- Cui, F.; Zhou, Z.; Zhou, H.S. Molecularly Imprinted Polymers and Surface Imprinted Polymers Based Electrochemical Biosensor for Infectious Diseases. Sensors 2020, 20, 996.
- Esfandyari-Manesh, M.; Javanbakht, M.; Shahmoradi, E.; Dinarvand, R.; Atyabi, F. The control of morphological and size properties of carbamazepine-imprinted microspheres and nanospheres under different synthesis conditions. J. Mater. Res. 2013, 28, 2677–2686.
- Holland, N.; Frisby, J.; Owens, E.; Hughes, H.; Duggan, P.; McLoughlin, P. The influence of polymer morphology on the performance of molecularly imprinted polymers. Polymer 2010, 51, 1578–1584.
- Rosengren, A.M.; Karlsson, B.C.G.; Nicholls, I.A. Consequences of Morphology on Molecularly Imprinted Polymer-Ligand Recognition. Int. J. Mol. Sci 2013, 14, 1207–1217.
- Pengkamta, T.; Mala, M.; Klakasikit, C.; Kanawuttikorn, P.; Boonkorn, P.; Chuaejedton, A.; Karuehanon, W. Synthesis and Evaluation of Molecularly Imprinted Polymer as a Selective Material for Vanillin. Suan Sunandha Sci. Technol. J. 2020, 7, 1–6.
- Xie, L.; Xiao, N.; Li, L.; Xie, X.; Li, Y. Theoretical Insight into the Interaction between Chloramphenicol and Functional Monomer (Methacrylic Acid) in Molecularly Imprinted Polymers. Int. J. Mol. Sci. 2020, 21, 4139.
- Dong, W.; Yan, M.; Liu, Z.; Wu, G.; Li, Y. Effects of solvents on the adsorption selectivity of molecularly imprinted polymers: Molecular simulation and experimental validation. Sep. Purif. Technol. 2007, 53, 183–188.
- Ansell, R.J. Characterization of the Binding Properties of Molecularly Imprinted Polymers. Adv. Biochem. Eng. Biotechnol. 2015, 150, 51–93.
- Shen, F.; Zhang, Q.; Ren, X. A triple-function zwitterion for preparing water compatible diclofenac imprinted polymers. Chem. Commun. 2015, 51, 183–186.
- Song, X.; Wang, J.; Zhu, J. Effect of Porogenic Solvent on Selective Performance of Molecularly Imprinted Polymer for Quercetin. Mater. Res. 2009, 12, 299–304.
- Hashim, S.N.N.S.; Boysen, R.I.; Schwarz, L.J.; Danylec, B.; Hearn, M.T.W. A comparison of covalent and non-covalent imprinting strategies for the synthesis of stigmasterol imprinted polymers. J. Chromatogr. A 2014, 1359, 35–43.
- Li, S.; Zhu, M.; Whitcombe, M.J.; Piletsky, S.A.; Turner, A.P.F. Molecularly Imprinted Polymers for Enzyme-like Catalysis: Principle, Design, and Applications. In Molecularly Imprinted Catalysts; Elsevier: Amsterdam, The Netherlands, 2016; pp. 1–17.
- Iacob, B.-C.; Bodoki, A.E.; Oprean, L.; Bodoki, E. Metal–Ligand Interactions in Molecular Imprinting. Ligand 2018, 23, 1875–1895.
- Bakhtiar, S.; Ahmad Bhawani, S.; Rizwan Shafqat, S. Synthesis and characterization of molecular imprinting polymer for the removal of 2-phenylphenol from spiked blood serum and river water. Chem. Biol. Technol. Agric. 2019, 6, 15.
- Yi, L.X.; Fang, R.; Chen, G.H. Molecularly imprinted solid-phase extraction in the analysis of agrochemicals. J. Chromatogr. Sci. 2013, 51, 608–618.
- Torres-Cartas, S.; Catalá-Icardo, M.; Meseguer-Lloret, S.; Simó-Alfonso, E.F.; Herrero-Martínez, J.M. Recent advances in molecularly imprinted membranes for sample treatment and separation. Separations 2020, 7, 69.
- BelBruno, P.P. Molecularly Imprinted Polymers. Chem. Rev. 2019, 119, 94–119.
- Sikiti, P.; Msagati, T.A.M.; Mamba, B.B.; Mishra, A.K. Synthesis and characterization of molecularly imprinted polymers for the remediation of PCBs and dioxins in aqueous environments. J. Environ. Health Sci. Eng. 2014, 12, 82.
- Bitas, D.; Samanidou, V. Molecularly imprinted polymers as extracting media for the chromatographic determination of antibiotics in milk. Molecules 2018, 23, 316.
- Xie, L.; Xiao, N.; Li, L.; Xie, X.; Li, Y. An investigation of the intermolecular interactions and recognition properties of molecular imprinted polymers for deltamethrin through computational strategies. Polymers 2019, 11, 1872.
- McStay, D.; Al-Obaidi, A.H.; Hoskins, R.; Quinn, P.J. Raman spectroscopy of molecular imprinted polymers. J. Opt. A Pure Appl. Opt. 2005, 7, s340–s345.
- Courtois, J.; Fischer, G.; Schauff, S.; Albert, K.; Irgum, K. Interactions of bupivacaine with a molecularly imprinted polymer in a monolithic format studied by NMR. Anal. Chem. 2006, 78, 580–584.
- Bompart, M.; De Wilde, Y.; Haupt, K. Chemical nanosensors based on composite molecularly imprinted polymer particles and surface-enhanced Raman scattering. Adv. Mater. 2010, 22, 2343–2348.
- Verheyen, E.; Schillemans, J.P.; Van Wijk, M.; Demeniex, M.A.; Hennink, W.E.; Van Nostrum, C.F. Challenges for the effective molecular imprinting of proteins. Biomaterials 2011, 32, 3008–3020.
