Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Cell Biology
Lactic acid bacteria (LAB) share several beneficial effects on human organisms, such as bioactive metabolites’ release, pathogens’ competition and immune stimulation.
- probiotic
- carrots
- Caenorhabditis elegans
1. Introduction
The Food and Agriculture Organization of the United Nations (FAO) and the World Health Organization (WHO) defined probiotics as live microorganisms that, when administered in adequate amounts, confer a health benefit on the host [1]. According to this statement, probiotics must be safe, and not exert pathogenic effects or show antibiotic resistance genes that could be transferred. Moreover, probiotic strains should be resistant to gastrointestinal conditions, such as stomach acid pH and bile acids, produce antimicrobial compounds and compete with pathogens by stimulating immunity [2]. Furthermore, probiotics’ efficacy should be confirmed in human studies.
Among the various microbial species associated with food, some of them may share probiotic features. The main source of probiotics used in humans is represented by dairy foods, but increasing evidence has highlighted the importance to select probiotics from other sources, such as fresh fruits and vegetables [3]. Indeed, the availability of commercial milk-based products limits their consumption by people who are intolerant or allergic to lactose. Therefore, fruits and vegetables offer healthy alternatives thanks to their large distribution and nutritive value. Among them, carrot (Daucus carota L.), as well as being rich in minerals and antioxidants, is reported to be a reservoir of carotenoids, vitamins and fiber [4][5][6]. Many studies on carrots have focused on cultivation, breeding, tissue culture, nutrient content and carotenoid synthesis regulation, while few works deal with microbial composition in terms of potential probiotic bacteria [7][8]. Indeed, the most common probiotics, isolated from fruits and vegetables, include different strains belonging to the lactic acid bacteria (LAB) group. This heterogeneous group of Gram-positive and non-spore-forming bacteria are normally present in food products, involved in numerous fermentation processes and some of them are widely used in industrial processes [9]. The major representatives of this microorganism group are Lactobacillus, Streptococcus, Leuconostoc, Pediococcus, Propionibacterium, Enterococcus, Bifidobacterium and Weissella genera [10]. Although the Weissella genus is found in multiple habitats, many species were isolated from different foods, such as fermented crop products, meat and fish, along with Leuconostoc species. Moreover, many of them produce exopolysaccharides, influencing the adhesion to substrates and affecting the structure of fermented foods.
Since the direct evaluation of probiotic potentials in vivo is often expensive and timewasting, the use of simple and inexpensive model systems is needed. Caenorhabditis elegans is a powerful in vivo model to screen for probiotic bacteria. Nematodes feed only on microorganisms, which reach the intestine, influencing nematodes’ physiology [11]. Among its many advantages, the possibility to easily monitor anti-aging markers or innate immunity pathways could be used for the screening of microorganisms to identify new probiotic strains and to explore the possible molecular pathways involved. Indeed, several foodborne LAB were reported to exert positive effects in worms, and the mechanisms correlated with innate immunity and lifespan extension have been elucidated [12]. Recently, different Lactobacillus strains, isolated from vegetables or dairy products, were reported to increase nematode viability, delay the aging process and protect against foodborne S. enterica serovar typhimurium LT2 or L. monocytogenes OH pathogens [13][14]. Moreover, it has been demonstrated that Bifidobacterium isolates can also exert beneficial effects on C. elegans health and lifespan [15].
2. Bacteria Isolation
In this study, potential probiotic properties of four bacterial isolates were evaluated in vitro and in vivo. Different bacterial colonies were isolated from carrots and identified at the molecular level by the amplification of 16S rDNA. The sequences obtained from sequencing were compared with those in the BLAST database, so that three strains belonging to Leuconostoc mesenteroides and a Weissella soli strain were identified. The Gram-positive strains isolated were identified as L. mesenteroides C1 (accession number OK513088), L. mesenteroides C2 (accession number OK513089), L. mesenteroides C7 (accession number OK513090) and Weissella soli T4 (accession number OK513091).
3. In Vitro Tests
3.1. Resistance to Lysozyme, Low pH and Bile Salts
To perform a selection of possible probiotic candidates, their resistance to the extreme conditions of the gastrointestinal tract, such as low pH in stomach and bile in the upper intestine, was evaluated. The high concentration of lysozyme present in the mouth represents the first barrier. Figure 1A reports bacteria survival data after 30 and 120 min of treatment with 1 mg/mL of lysozyme. All LAB strains showed high resistance to lysozyme after 30 min of incubation, with percentages of survival of 100%. Notably, L. mesenteroides C2 was able to resist even after 120 min of incubation with lysozyme, which can be considered a severe treatment. On the other hand, W. soli T4 and L. mesenteroides C7 showed a percent survival of 60% and 80% respectively, after 2 h of incubation. L. mesenteroides C1, instead, did not resist lysozyme treatment. Tolerance to low pH conditions was performed to simulate microbial flux along the mammalian gastrointestinal tract. As shown in Figure 1B, 3 h of incubation in pH 2.5 exerted a strong reduction (about 80%) of L. mesenteroides C1, L. mesenteroides C7 and W. soli T4 counts, as compared to the control. This result suggested that these strains were not able to endure acidic environments. Notably, L. mesenteroides C2 cell recovery showed a higher ability to survive in low pH conditions, similarly to the probiotic LGG strain. Similar results were obtained after testing the ability of different strains to resist at pH 3.0.
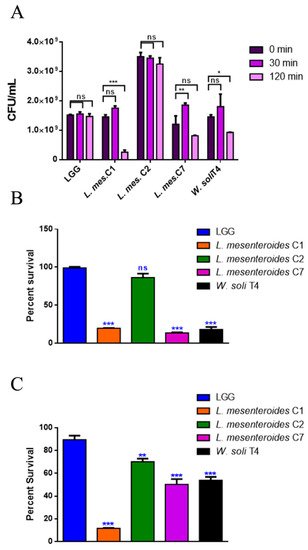
Figure 1. In vitro tolerance to lysozyme, pH 2.5 and 0.3% bile salts. (A) Cell counts of viable bacteria recovered at the initial time point (t0), following 30 or 120 min of incubation in 1 mg/mL of lysozyme SES buffer. (B) Recovery of viable bacteria after 3 h of incubation in phosphate buffer adjusted to pH 2.5 or (C) 0.3% bovine bile salts. LGG was taken as the LAB reference strain. Columns represent the mean ± SD of three independent experiments. Statistical analysis was performed by one-way ANOVA, followed by the Bonferroni post-test. Asterisks indicate significant differences (* p < 0.05; ** p < 0.01; *** p < 0.001), ns: not significant.
The human bile concentration ranges from 0.3% to 0.5%, and a high bile tolerance improves probiotics’ colonization in the host GI tract [16]. To investigate the ability of the different isolates to survive in the presence of bile, the percent survival in the presence of 0.3% bile was tested. As shown in Figure 1C, L. mesenteroides C2, L. mesenteroides C7 and W. soli T4 strains showed a growth percentage above 50% in the presence of bile. As expected, the reference probiotic strain LGG exhibited a resistance of about 90%. On the other hand, L. mesenteroides C1 was not able to resist the treatment, showing a decrease in viability of about 90%.
3.2. Antibiotic Susceptibility and Antagonistic Activity to Pathogens
Antibiotic susceptibility was determined by the disc diffusion assay, analyzing a panel of 20 antibiotics. Among them, there are inhibitors of synthesis of cell wall, DNA and RNA, proteins and inhibitors of membrane function. As reported in Table 1, LAB isolates showed an antibiotic susceptibility pattern very similar to that of the LGG control strain. L. mesenteroides C2 displayed resistance to only five antibiotics, and in some cases, the inhibition halo was larger as compared to the control (Table 2).
Table 1. Primers for real-time qPCR analysis.
| hsf-1 | FOR | 5′-ATGACTCCACTGTCCCAAGG |
| REV | 5′-TCTTGCCGATTGCTTTCTCT | |
| pmk-1 | FOR | 5′-AAATGACTCGCCGTGATTTC |
| REV | 5′-CATCGTGATAAGCAGCCAGA | |
| sod-3 | FOR | 5′-AGAACCTTCAAAGGAGCTGATG |
| REV | 5′-CCGCAATAGTGATGTCAGAAAG | |
| act-1 | FOR | 5′-GAGCGTGGTTACTCTTTCA |
| REV | 5′-CAGAGCTTCTCCTTGATGTC | |
| skn-1 | FOR | 5′-GTTCCCAACATCCAACTACG |
| REV | 5′-TGGAGTCTGACCAGTGGATT | |
| daf-16 | FOR | 5′-TCAAGACCTCAAAGCCAATCAACTC |
| REV | 5′-ACGAGAAAGAAGGAGTAAGAGGAGG |
Table 2. Resistance to antibiotics of different isolates. The zones of inhibition were measured from the center of the disc and recorded in mm ± SD. Absence of inhibition halo was indicated as (+), ns: not significant.
| Antibiotic | Amount on Disc (µg) | Zone of Inhibition (mm) | |||||
|---|---|---|---|---|---|---|---|
| LGG | L. mesenteroides C1 | L. mesenteroides C2 | L. mesenteroides C7 | W. soli T4 | p-Value | ||
| Amikacin | 30 | 4 ± 0.03 | 5 ± 0.06 | 10 ± 0.08 | 4 ± 0.05 | 4 ± 0.03 | p < 0.001 |
| Ampicillin | 10 | 7 ± 0.06 | 7 ± 0.05 | 6 ± 0.10 | 5 ± 0.20 | 5 ± 0.05 | p < 0.01 |
| Aztreonam | 30 | + | + | + | + | + | ns |
| Carbenicillin | 100 | 8 ± 0.08 | 7 ± 0.03 | 5 ± 0.09 | 7 ± 0.03 | 5 ± 0.08 | p < 0.01 |
| Cefalotin | 30 | + | 3 ± 0.10 | + | 4 ± 0.08 | + | p < 0.01 |
| Cefotaxime | 30 | 7 ± 0.08 | + | 7 ± 0.20 | + | + | p < 0.001 |
| Cefuroxime | 30 | 5 ± 0.02 | + | 12 ± 0.08 | + | + | p < 0.001 |
| Clindamycin | 2 | 8 ± 0.12 | 9 ± 0.35 | 4 ± 0.08 | 10 ± 0.10 | 9 ± 0.30 | p < 0.01 |
| Chloramphenicol | 30 | 8 ± 0.08 | 8 ± 0.35 | 14 ± 0.10 | 8 ± 0.20 | 10 ± 0.15 | p < 0.01 |
| Erythromycin | 15 | 8 ± 0.03 | 7 ± 0.08 | 8 ± 0.15 | 7 ± 0.09 | 8 ± 0.08 | ns |
| Fosfomycin | 50 | + | + | + | + | + | ns |
| Gentamicin | 10 | 4 ± 0.05 | 5 ± 0.12 | + | 5 ± 0.15 | 5 ± 0.10 | p < 0.05 |
| Mezlocillin | 75 | 14 ± 0.05 | 10 ± 0.15 | 13 ± 0.20 | 8 ± 0.20 | 11 ± 0.10 | p < 0.001 |
| Oxacillin | 1 | + | + | 4 ± 0.03 | + | + | p < 0.05 |
| Penicillin | 10 | 13 ± 0.10 | 9 ± 0.20 | 14 ± 0.20 | 9 ± 0.05 | 11 ± 0.09 | p < 0.001 |
| Rinfampicin | 30 | 13 ± 0.02 | 11 ± 0.09 | 8 ± 0.10 | 11 ± 0.08 | 10 ± 0.08 | p < 0.01 |
| Streptomycin | 25 | 4 ± 0.12 | 5 ± 0.08 | + | 4 ± 0.06 | 4 ± 0.05 | p < 0.01 |
| Tetracycline | 30 | 14 ± 0.15 | 10 ± 0.30 | 7 ± 0.05 | 8 ± 0.06 | 7 ± 0.10 | p < 0.001 |
| Tobramycin | 10 | 4 ± 0.08 | 4 ± 0.05 | 4 ± 0.09 | 3 ± 0.05 | 4 ± 0.05 | ns |
| Vancomycin | 30 | + | + | + | + | + | ns |
The antagonistic activity was evaluated through the agar double-layer diffusion test, against four pathogen test strains: the Gram-positive Staphylococcus aureus and Listeria monocytogenes, and the Gram-negative Pseudomonas aeruginosa and S. enterica serovar typhimurium LT2. The antagonistic activity was less variable among the different isolates, as shown by inhibition halo diameters on all four pathogen test strains (Table 3). Indeed, the inhibition halo diameters produced by the different isolates were comparable to that of the probiotic strain LGG.
Table 3. Antagonistic activity in vitro. The diameter of inhibition halos was recorded in mm and the data were expressed as average ± SD.
| Pathogen | LGG | L. mesenteroides C1 | L. mesenteroides C2 | L. mesenteroides C7 | W. soli T4 | p-Value |
|---|---|---|---|---|---|---|
| S. aureus | 40 ± 0.08 | 38 ± 0.4 | 35 ± 0.5 | 35 ± 0.1 | 35 ± 0.5 | p < 0.01 |
| L. monocytogenes | 30 ± 0.2 | 29 ± 0.2 | 30 ± 0.8 | 31 ± 0.6 | 30 ± 0.5 | ns |
| P. aeruginosa | 40 ± 0.07 | 38 ± 0.5 | 33 ± 0.5 | 40 ± 0.1 | 35 ± 0.6 | p < 0.01 |
| S. enterica | 30 ± 0.3 | 31 ± 0.3 | 30 ± 0.5 | 28 ± 0.5 | 30 ± 0.08 | ns |
This entry is adapted from the peer-reviewed paper 10.3390/microorganisms9112290
References
- FAO/WHO Probiotics in Food. Health and Nutritional Properties and Guidelines for Evaluation 2006. Available online: http://www.fao.org/food/food-safety-quality/a-z-index/probiotics/en/ (accessed on 16 September 2021).
- Markowiak-Kopeć, P.; Śliżewska, K. Effects of Probiotics, Prebiotics, and Synbiotics on Human Health. Nutrients 2017, 9, 1021.
- Roselli, M.; Natella, F.; Zinno, P.; Guantario, B.; Canali, R.; Schifano, E.; De Angelis, M.; Nikoloudaki, O.; Gobbetti, M.; Perozzi, G.; et al. Colonization Ability and Impact on Human Gut Microbiota of Foodborne Microbes from Traditional or Probiotic-Added Fermented Foods: A Systematic Review. Front. Nutr. 2021, 8, 689084.
- Sciubba, F.; Tomassini, A.; Giorgi, G.; Brasili, E.; Pasqua, G.; Capuani, G.; Aureli, W.; Miccheli, A. NMR-Based Metabolomic Study of Purple Carrot Optimal Harvest Time for Utilization as a Source of Bioactive Compounds. Appl. Sci. 2020, 10, 8493.
- Tomassini, A.; Sciubba, F.; Di Cocco, M.E.; Capuani, G.; Delfini, M.; Aureli, W.; Miccheli, A. 1H NMR-Based Metabolomics Reveals a Pedoclimatic Metabolic Imprinting in Ready-to-Drink Carrot Juices. J. Agric. Food Chem. 2016, 64, 5284–5291.
- Arscott, S.A.; Tanumihardjo, S.A. Carrots of Many Colors Provide Basic Nutrition and Bioavailable Phytochemicals Acting as a Functional Food. Compr. Rev. Food Sci. Food Saf. 2010, 9, 223–239.
- Que, F.; Hou, X.-L.; Wang, G.-L.; Xu, Z.-S.; Tan, G.-F.; Li, T.; Wang, Y.-H.; Khadr, A.; Xiong, A.-S. Advances in research on the carrot, an important root vegetable in the Apiaceae family. Hortic. Res. 2019, 6, 69.
- Da Silva, E.A.; Vieira, M.A.; Vieira, E.A.; Amboni, R.D.D.M.C.; Amante, E.R.; Teixeira, E. Chemical, Physical and Sensory Parameters of Different Carrot Varieties (Daucus carota L.). J. Food Process. Eng. 2007, 30, 746–756.
- Forssten, S.D.; Sindelar, C.W.; Ouwehand, A. Probiotics from an industrial perspective. Anaerobe 2011, 17, 410–413.
- Sornplang, P.; Piyadeatsoontorn, S. Probiotic isolates from unconventional sources: A review. J. Anim. Sci. Technol. 2016, 58, 26.
- Poupet, C.; Chassard, C.; Nivoliez, A.; Bornes, S. Caenorhabditis elegans, a Host to Investigate the Probiotic Properties of Beneficial Microorganisms. Front. Nutr. 2020, 7, 135.
- Kwon, G.; Lee, J.; Lim, Y.-H. Dairy Propionibacterium extends the mean lifespan of Caenorhabditis elegans via activation of the innate immune system. Sci. Rep. 2016, 6, 31713.
- Schifano, E.; Zinno, P.; Guantario, B.; Roselli, M.; Marcoccia, S.; Devirgiliis, C.; Uccelletti, D. The Foodborne Strain Lactobacillus fermentum MBC2 Triggers pept-1-Dependent Pro-Longevity Effects in Caenorhabditis elegans. Microorganisms 2019, 7, 45.
- Guantario, B.; Zinno, P.; Schifano, E.; Roselli, M.; Perozzi, G.; Palleschi, C.; Uccelletti, D.; Devirgiliis, C. In Vitro and In Vivo Selection of Potentially Probiotic Lactobacilli from Nocellara del Belice Table Olives. Front. Microbiol. 2018, 9, 595.
- Roselli, M.; Schifano, E.; Guantario, B.; Zinno, P.; Uccelletti, D.; Devirgiliis, C. Caenorhabditis Elegans and Probiotics Interactions from a Prolongevity Perspective. Int. J. Mol. Sci. 2019, 20, 5020.
- Dunne, C.; O’Mahony, L.; Murphy, L.; Thornton, G.; Morrissey, D.; O’Halloran, S.; Feeney, M.; Flynn, S.; Fitzgerald, G.; Daly, C.; et al. In vitro selection criteria for probiotic bacteria of human origin: Correlation with in vivo findings. Am. J. Clin. Nutr. 2001, 73, 386s–392s.
This entry is offline, you can click here to edit this entry!
