Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
The gastrointestinal tract is the body’s largest interface between the host and the external environment. People infected with SARS-CoV-2 are at higher risk of microbiome alterations and severe diseases. Recent evidence has suggested that the pathophysiological and molecular mechanisms associated with gastrointestinal complicity in SARS-CoV-2 infection could be explained by the role of angiotensin-converting enzyme-2 (ACE2) cell receptors.
- gut flora
- neurological symptoms
- coronavirus disease 19
1. Introduction
Coronavirus disease 2019 (COVID-19) is an infectious respiratory disease caused by a new coronavirus—namely, SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) [1]. As of 23 August 2021 [2], it has infected over 211 millions of people in more than 190 countries and caused more than 4.7 million deaths all over the world. In order to control and manage the COVID-19 pandemic and avoid the recurrence of future pandemics, the world’s scientists have attempted to find appropriate solutions through interdisciplinary cooperation [3]. During the first decade of the 21st century, development in life sciences has provided us with a deep understanding of the interactions between pathogens and their hosts. For example, there have been remarkable innovations in molecular biology in discovering pathogenesis mechanisms and diagnosis techniques, as well as advances in drug discovery, vaccines, and computational simulation due to the occurrence of various epidemics [4][5].
Many researchers have presented valuable information on the underlying events of SARS-CoV-2 infection and its related symptoms. Although COVID-19 is mainly a pulmonary disease, gastrointestinal symptoms have also been reported in some patients with this virus [6]. Respiratory symptoms, fever, and cough, as the main clinical signs among COVID-19 patients, have been investigated more than other symptoms, and general agreement about others, including the outbreak of gastrointestinal (GI) symptoms, is lacking [7].
Results indicate that the SARS-CoV-2 can damage the gastrointestinal system directly or indirectly through inflammatory reactions and may lead to several digestive problems, including vomiting, diarrhea, nausea, diminished appetite, and abdominal pain in patients with COVID-19 [8][9]. It has been determined that these gastrointestinal symptoms along with intestinal flora dysbiosis occur due to SARS-CoV-2 attacking the digestive tract of the host. Based on some reports, it seems that individuals with gastrointestinal problems are more likely to experience severe COVID-19 disease, which may be seen as a predictor of the development of severe respiratory disorders [10][11]. Additionally, several reports and pieces of evidence suggest that SARS-CoV-2 most likely either directly or indirectly affects the enteric nervous system, leading to gut dysfunction and neuro-gastrointestinal manifestations [12].
The hepatic consequences of SARS-CoV-2 infection are an important problematic component of COVID-19 that is most important in patients with earlier liver disease who are at remarkably high risk of severe COVID-19 and death [13][14][15]. Although the entire impact of COVID-19 on the liver is not clear, biochemical abnormalities in the liver are occurring in approximately 15–65% of people infected with SARS-CoV-2. Some reports have demonstrated that an elevation in serum liver enzymes is associated with adverse outcomes such as shock, intensive care unit (ICU) admission, and mechanical ventilation. These findings were made by Marjot et al. [16] in a review article. Additionally, the possible underlying direct and indirect mechanisms of liver injury have also been discussed.
2. Gastrointestinal Pathogenesis of SARS-CoV-2
Based on the reports, SARS-CoV-2 shares common clinical and epidemiological features with SARS-CoV and MERS CoV (Middle East Respiratory Syndrome coronavirus) due to its 70% and 40% similarity, respectively, in genetic sequence [17]. It is well established that angiotensin converting enzyme-2 (ACE2), a relevant player in the renin-angiotensin system (RAS), is the key receptor for SARS-CoV-2 invasion and the infection of human target cells that is similar to SARS-CoV. ACE2 converts angiotensin-2 to angiotensin-(1–7), which negatively regulates the active RAS [18]. Previous investigations of SARS-CoV have shown that its cellular entry occurs via the epithelial ACE2 receptor [19]. For this purpose, the receptor-binding domain (RBD) of viral surface glycoproteins named the spike (S) protein binds to the ACE2 receptor with a high affinity and thus releases the virus to the cell [20]. ACE2, a type I transmembrane glycoprotein, consists of a longer extracellular area with carboxypeptidase activity and a short intracellular cytoplasmic tail [21].
The trimeric spike (S) glycoproteins on the viral surface are composed of about 1300 amino acid residues and contain three main parts: the extracellular domain, the transmembrane domain, and the intracellular tail. The spike protein is cleaved by host cell transmembrane serine protease 2 (TMPRSS2) into S1 (the peripheral fragment) and S2 (membrane-spanning fragment) subunits. The S1 subunit contains a receptor-binding domain with several disordered regions, facilitating interactions with host cell receptors, whereas the S2 subunit fuses with the host cell membrane and releases the viral RNA for replication and translation by the host cell machinery [22][23][24].
The interaction of ACE-2 with spike proteins has been presented previously in a comprehensive review. Additionally, the small molecular inhibitors that can limit these interactions and be used as potential therapeutic platforms have been discussed [25].
Researchers have employed single-cell sequencing to identify the amount of ACE2 expressed in several organs. The obtained results show that ACE2 was largely found not only in the lung tissue but that it is also overexpressed in the gastrointestinal tract, including the esophagus and the absorptive intestinal epithelium in the colon and ileum. The ACE2 expression in the gastrointestinal tract, particularly in the colon, is approximately 100-fold higher than that in the alveolar cells of the lungs and respiratory system [26][27].
In another study, results from ACE2 immunohistochemical staining showed that the liver and digestive organs have higher ACE2 expression levels compared to the respiratory system. However, in lung tissue, the ACE2 expression level increased with age, which might explain, to some extent, why elderly people with COVID-19 are more likely to develop pneumonia. Recently, research on the pathophysiology of COVID-19 has indicated that the pattern and level of human ACE2 enzyme expression in different tissues might be correlated with the various different symptoms and outcomes of COVID-19 [28]. The abundance of this receptor in the epithelium of the GI tract and digestive system, especially in the small and large intestines, makes them susceptible to virus entry.
Some symptoms in patients with COVID-19, such as cardiovascular, kidney, GI, and brain manifestations, are associated with the co-expression of ACE2 and TMPRSS2. The intestinal brush border cells have high levels of TMPRSS2, transmembrane serine protease 2, which is capable of robust and persistent infection. These cells would be a useful tool for understanding the pathogenesis of SARS-CoV-2 in the GI tract [14]. TMPRSS2 and ACE-2 co-expression in GI could be the entry route for SARS-CoV-2 virus in absorptive enterocytes of the colon and ileum, resulting in further damage to the mucous membrane barrier, the development of inflammatory cytokine production, and the GI symptoms associated with COVID-19. Expression of ACE2 can be altered by several factors, including GI malignancies, which could also increase the severity of COVID-19 infection due to higher expression of ACE-2 and TMPRSS2 [29]. These findings suggest that a high expression of ACE2 in the digestive system may be a potential route of infection. Therefore, in COVID-19 patients, understanding the mechanism of damage to the digestive tract is very important in order to guide clinical treatment.
Perisetti et al. have reported that people with hypochlorhydria are more sensitive to viral infections due to a higher viral load entering the small intestine via ACE2 [30]. Sung et al. concluded that the down regulation of ACE2 by SARS-CoV-2 changes the uptake of certain amino acids; disrupts the gut barrier; increases the lipopolysaccharide and peptidoglycan levels of bacteria; and promotes systemic inflammation, contributing to the occurrence of a cytokine storm [31]. Based on these findings, the mechanism correlated with GI tract manifestations in SARS-CoV-2 infection could be further studied through the mediation of ACE2 receptors.
2.1. Microbiome Alterations, a Putative Mechanism of Gastrointestinal Manifestations in COVID-19 Patients
Normal intestinal flora (referring to the collective genomes of microorganisms including bacteria, fungi, viruses, etc.) plays a crucial role in maintaining the homeostasis of the immune system and the metabolic function and health of the host gastrointestinal tract. Normal human gut flora harbors trillions of microbes that play important roles in human biology and disease. A change in the most abundant genera of the intestinal flora may result in some pathological disorders, such as GI dysfunction, including diarrhea, nausea, and vomiting; inflammatory bowel disease (IBD); and neurodegenerative problems. According to Martinez-Guryn et al., much about this topic remains to be revealed, including distinguishing healthy versus unhealthy gut microbiomes, which are likely to be characterized by the host, pathogens, and environmental factors [32].
It has been shown that COVID-19 infection increases the risk of gut flora changes in patients. Therefore, it should be expected that the digestive disorders and the immune homeostasis alterations induced by the virus might be mediated, to some extent, by the gut flora [33][34]. Some patients with COVID-19 have shown a gut microbial imbalance, with low levels of Bifidobacterium and Lactobacillus [35]. It has been reported that the amounts of Clostridium hathewayi, Coprobacillus, and Clostridium ramosum, present are closely correlated with the severity of COVID-19 and inversely correlated with Faecalibacterium prausnitzii [36]. Furthermore, antimicrobials significantly alter the gut microbiota pattern, resulting in Clostridioides difficile infection as a consequence of bacterial disturbances in the gut. The gut microbiota plays an essential role in opposing the colonization of Clostridioides difficile, and this microbial perturbation creates a favorable environment for colonization and Clostridioides difficile infection [37][38]. Therefore, clinicians should be aware of the disadvantages of the extensive use of antibiotics and, consequently, possible Clostridioides difficile infection with SARS-CoV-2 co-infection.
Generally, several key factors in microbiome alterations in people infected with SARS-CoV-2 have been highlighted by Perisetti et al. [30]. The first is the increase in the release of pro-inflammatory cytokines. The second is antimicrobial medications, including antibiotics and antivirals. The third is changes in the lung flora and the ratio of pathogenic organisms. The fourth is enteral nutrition. Additionally, the fifth is aberrant mTOR (mechanistic Target of Rapamycin Kinase) activity [30]. All of these mechanisms are only potential, and there is no credible proof of whether they act separately or in collaboration with each other in the progression of digestive symptoms in COVID-19 patients [30]. Some researchers have suggested that the targeting of the intestinal flora could potentially be a useful strategy with which to fight SARS-CoV-2 infection [33].
2.1.1. Pro-Inflammatory Cytokines
It is well known that patients infected with SARS-CoV-2 show increased levels of cytokines and inflammation biomarkers [30]. In fact, COVID-19 disease leads to an excessive activated immune response, with the uncontrolled production and release of a number of cytokines. This phenomenon, known as a cytokine storm, is the main reason for worsening of the condition of COVID-19 patients, including those with gastrointestinal disorders, which alters the gut motility and GI flora. A cytokine—namely, granulocyte monocyte colony-stimulating factor (GM-CSF)—is made up of different types of cells, such as macrophages, T cells, fibroblasts, and endothelial cells, and is heavily loaded in the gastrointestinal tract [39].
The first line of host defense against attacking pathogens relies on pattern recognition receptor (PRR)-mediated signaling (mostly Toll-like receptors) [40]. RNA release is recognized by RIG-I (viral RNA receptor retinoic-acid inducible gene I), cytosolic MDA5 receptor (melanoma differentiation-associated gene5), STING (stimulator of interferon genes), and cGA Snucleotidyltransferase (cyclic GMP-AMP synthase). This leads to the activation of downstream signaling, including pro-inflammatory factors (e.g., IL-6), antiviral cytokines, NF-κB (nuclear factor-κB), and IFN (interferon) [41][42]. COVID-19 patients show a suppressed Nrf2 (nuclear factor erythroid 2-related factor 2) pathway. Nrf2 is a cytoprotective factor that inhibits NF-κB and inhibits the expression of the inflammatory cytokines in macrophages in SARS-CoV-2-infected cells. Nrf2 reduces the ACE2 receptor expression in respiratory epithelial cells. Nrf2 activation plays a role in the execution of inflammation and decreases the intensity of cytokine storms [43]. Additionally, high levels of IL-6 play a key role in worsening cytokine storms, caused by respiratory failure and acute respiratory distress syndrome. IL-6 mainly makes use of two pathways: cis and trans (Figure 1). The cis pathway is important for the regenerative and protective functions of IL-6. In this pathway, the binding of IL-6 to its receptor, mIL-6R, and gp130 leads to the activation of the JAK (Janus kinase)/STAT3 (signal transducer and activator of transcription 3) pathway. Then, innate and acquired immunity is activated by this pathway, which leads to cytokine release syndrome. The trans pathway is also responsible for the pro-inflammatory activity of the cytokine [44]. Hence, Nrf2 and IL-6 may be introduced as therapeutic targets for COVID-19, especially for patients suffering from inflammatory problems. However, some trials have reported a lack of improvement in COVID-19 patient mortality upon IL-6 inhibitor treatment. According to Lowery et al., the most important reason for this paradox is that the pathogenic mechanisms of this viral disease are similar but not the same in all patients [45].
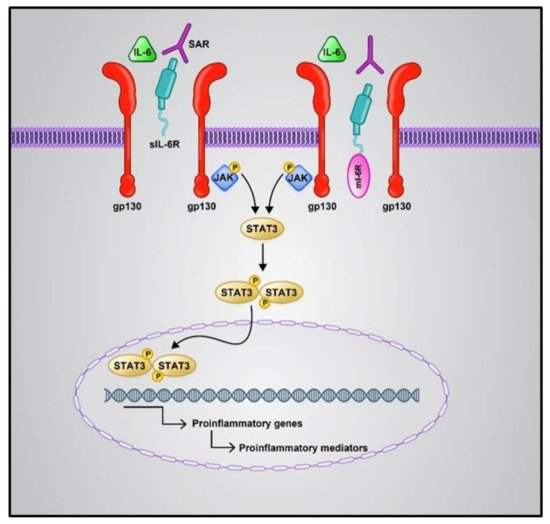
Figure 1. Cis and trans IL6 signaling pathways. The cis pathway is mediated by mIL6R as an anti-inflammatory, while the trans-signaling pathway is mediated by sIL6-R as a pro-inflammatory. Both the IL-6 signaling pathways converge in the activation of the JAK/STAT pathway. Phosphorylation of the receptor associated JAK1 is induced by the binding of numerous cytokines or growth factors to their specific receptors; then, STAT3 phosphorylation occurs via JAK1. Phosphorylated STAT3 is dimerized and transferred to the nucleus, which causes the expression of target genes contributing to angiogenesis, proliferation, immunosuppression, and inflammation; IL6: Interlukine 6, mIL6R: membrane form of IL-6 receptor, sIL6-R: soluble form of IL-6 receptor, JAK: Janus kinase, STAT: signal transducer and activator of transcription.
It should be noticed that COVID-19 patients in earlier stages of inflammatory diseases and GI symptoms, including malabsorption syndromes and IBD, are at a high risk of aggravating GI manifestation. It has been reported that the level of fecal calprotectin, a marker of bowel inflammation, is elevated in COVID-19 patients who suffer from diarrhea for more than 48 h [30]. It was found that ACE2 is highly expressed in inflammatory states, especially in IBD [46]. In IBD, age, inflammation, and disease location were identified as acute factors of the intestinal expression of ACE2. The role of ACE2 is controversial. On the one hand, it can regulate gut inflammation and diarrhea. However, on the other hand, its interaction with SARS-CoV-2 could lead to diarrhea [47]. Furthermore, it is known that ACE2 plays a significant role in gastric ulcer healing, which can be correlated with virus-mediated diarrhea [48]. Therefore, to provide a deep understanding of the aggravating symptoms in COVID-19 patients, especially those with gastric disorders, and determine therapeutic targets, ACE2-related signaling needs to be studied further in these patients.
2.1.2. Antimicrobial Medications
Based on the available evidence, the use of different antimicrobial drugs such as antibiotics and antivirals can alter the gut flora and sensitize people to adverse GI effects [30]. Recent studies suggest that patients with altered gut microbiota might experience more severe COVID-19 symptoms. Diarrhea, nausea, and stomach pain are common adverse effects that can be induced by the various antivirals and antibiotics that are used in COVID-19 patient management [49]. However, through further research, whether these adverse effects have diagnostic value for COVID-19 could be determined [47]. Given that the use of antibiotic medications in COVID-19 patients could further change the digestive microbial flora, bacterial pneumonia treatment should only be initiated when clinical suspicion is high. However, the best antibiotic to use in COVID-19 patients with bacterial co-infections and GI symptoms remains unclear. In a review article published by Chedid et al. [48], data from 19 studies regarding antibiotic consumption in 2834 COVID-19 patients are analyzed. The use of antibiotic drugs occurred in 74% of patients. Only 17.6% of patients with COVID-19 who received antibiotics had secondary infections and 50% of the patients who received antibiotics were not severely ill, indicating the significant desire to initiate the use of antibiotics in mildly or moderately ill patients. Some retrospective studies have reported that antibiotic medication usage had no clear positive effect on mortality. However, detailed information on antibiotic treatment is lacking in most studies. Additional research to determine related manifestations for antibiotic use in COVID-19 patients is critical in light of the significant level of mortality associated with secondary infections in these patients, and the rising rate of antimicrobial resistance [50]. Some antivirals and antibiotics that have been used in COVID-19 treatment and reported on by several authors are listed in Table 1.
Table 1. Some antiviral and antibiotics that are used in COVID-19 treatment, as reported by Chedid 2021 [50], Bagheri 2021 [51], and Frediansyah 2021 [52].
| Drug/Drug Class | Therapeutic Category | Reference |
|---|---|---|
| Aminoglycosides | Antibiotic | Bagheri 2021 |
| Azithromycin | Antibiotic | Bagheri 2021, Chedid 2021 |
| Moxifloxacin | Antibiotic | Chedid 2021 |
| Ceftriaxone | Antibiotic | Chedid 2021 |
| Cephalosporin | Antibiotic | Chedid 2021 |
| Quinolones | Antibiotic | Chedid 2021 |
| Clarithromycin | Antibiotic | Chedid 2021 |
| Ceftriaxone | Antibiotic | Chedid 2021 |
| Tigecycline | Antibiotic | Chedid 2021 |
| Cefoperazone | Antibiotic | Chedid 2021 |
| Umifenovir | Antiviral | Frediansyah 2021 |
| Lopinavir | Antiviral | Frediansyah 2021 |
| Darunavir | Antiviral | Frediansyah 2021 |
| Atazanavir | Antiviral | Frediansyah 2021 |
| Saquinavir | Antiviral | Frediansyah 2021 |
| Emtricitabine | Antiviral | Frediansyah 2021 |
| Azvudine | Antiviral | Frediansyah 2021 |
| Remdesivir | Antiviral | Frediansyah 2021 |
| Favipiravir | Antiviral | Frediansyah 2021 |
| Ribavirin | Antiviral | Frediansyah 2021 |
| Sofosbuvir | Antiviral | Frediansyah 2021 |
| Oseltamivir | Antiviral | Frediansyah 2021 |
2.1.3. Lung Flora Changes and Changes in the Ratio of Pathogenic Organisms
Based on recent reports on patients with respiratory problems, the “gut–lung” axis has been identified as a possible cause of GI disorders. Microbiota metabolites and the gut microbiota can regulate lung immunity through the lymphatic or circulatory systems. Several studies have reported a correlation between intestinal flora changes and disease exacerbation, and changes in the gut flora have been linked to lung disorders and infections of the respiratory tract [53][54][55]. Similarly, lung flora changes due to respiratory disorders in COVID-19 patients can be affected by the composition of the gut flora [30]. Immune reactions of the intestinal mucosal barrier, which protects the host against thousands of pathogens and environmental antigens, seem to correlate with pulmonary immune reactions [56][57][58] (Figure 2).
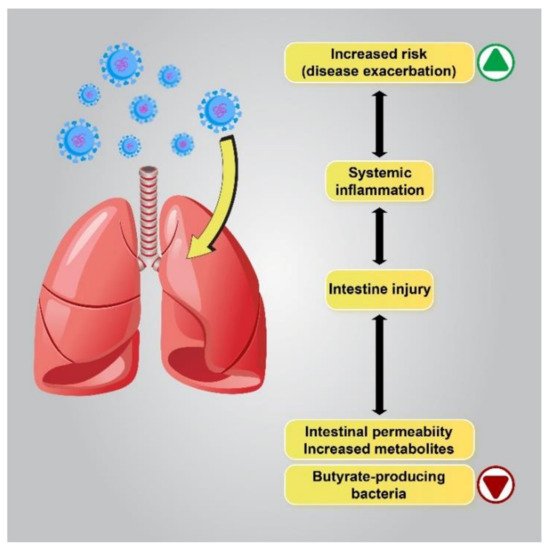
Figure 2. The correlation between flora changes and disease exacerbation. Patients with a lower gut abundance of butyrate-producing bacteria are more likely to develop viral lower respiratory tract infections. Dysbiosis alters metabolite production in the gut by enhancing the permeability of the intestines, which leads to the intensification of pre-existing lung diseases or an increase in the risk of respiratory diseases.
Disrupted gut barrier integrity related to senility or underlying chronic disease, including obesity, diabetes, and hypertension, may be a main factor that permits the virus to gain access to the ACE2 receptor on the enterocytes and leak out of the digestive system to spread throughout the body. Therefore, if the gut immune barrier is disrupted, attacking microorganisms can enter the bloodstream or lungs and cause septicemia and acute respiratory distress syndrome (ARDS) [59]. Interestingly, several studies have demonstrated a close connection between COVID-19 severity and gut microbiota dysbiosis to be common. It has been reported that the abundance of beneficial bacteria belonging to the families Ruminococcaceae or Lachnospiraceae, the species Faecalibacterium prausnitzii, and the class Clostridia was reduced in COVID-19 patients. The class Clostridia is one of the major butyric acid-producing bacteria in the gut. The penetration of SARS-CoV-2 into the gut barrier may cause inflammation due to excessive immune responses that further increase gut permeability. In contrast, in the GI tract of healthy people or young children, who have a higher number of regulatory T cells (Treg cells) due to their activation by butyrate, the virus may be contained in the digestive system without posing a considerable threat to the other organs of the body, eventually being excreted in the feces. Therefore, further investigation of the complex microbial interactions involving important butyrate-producing species is essential in order to understand its influence on human health and disease [60][61].
In a study conducted by Bradley et al., it was found that an abundance of segmented filamentous bacteria provokes the migration of Th17 cells to the lung, enhancing the autoimmune response and worsening pulmonary lesions [62].
Changes in the number of pathogens is another factor of microbiome alterations in people infected with SARS-CoV-2. Furthermore, altered flora also affects the ratio of pathogenic organisms, potentially leading to other infections such as Clostridioides difficile occurring in COVID-19 patients [30]. Foreign pathogenic organisms after infection with the virus and due to enterocyte absorption disorders increase the permeability of the intestinal barrier. Hence, theoretically, intestinal symptoms (such as diarrhea) demonstrate that the digestive system may be sensitive to SARS-CoV-2 infection [27].
2.1.4. Enteral Nutrition
Given that no specific antiviral medications are available for COVID-19 patients, the immune strength of patients is crucial. Nutrition support is necessary to achieve adequate immune function. Oral feeding is a priority. When oral nutrition is difficult, enteral feeding should be considered as early as possible. In patients with acidosis, uncontrolled bleeding of the GI, uncontrolled hypoxemia, or other harmful conditions, delayed enteral feeding is suggested. Enteral feeding has been proposed for the maintenance of normal intestinal mucosal barrier function, as it prevents intestinal microbiome translocation and reduces the occurrence of infectious complications [63][64]. Enteral nutrition usage might alter the gut flora, which plays a significant role in maintaining GI homeostasis [30].
2.1.5. Aberrant mTOR Activity and Deficit of ACE2
ACE-2 has been identified as a key regulator of neutral amino acid transporter in the small intestine and its deficit leads to the critical deficiency of local tryptophan homeostasis, which could alter the intestinal microbiome and an individual’s susceptibility to inflammation [65]. Consequently, impaired amino acid adsorption can lead to a low expression of antimicrobial peptides, leading to a change in the composition of the gut microbiota. ACE2 is essential for the surface expression of B0AT1 (the neutral amino acid transporter) in the small intestine. Dietary tryptophan is primarily absorbed via the transport pathway of B0AT1/ACE2 in the epithelial cells of the small intestinal; this results in mTOR pathway activation and the regulation of the expression of antimicrobial peptides. These peptides can adjust the gut microbiota composition [66]. The blocking of ACE2 and aberrant mTOR activity after SARS-COV-2 infection can molecularly explain how amino acid malnutrition results in intestinal inflammation and diarrhea. As SARS-CoV-2 S protein binds to the ACE2 receptor, blocking ACE2 leads to B0AT1 being blocked and thus tryptophan absorption being disturbed, leading to the alteration of the microbiota due to the aberrant secretion of antimicrobial peptides [66]. Hence, aberrant mTOR activity leads to a decreased expression of antimicrobial peptides from small intestinal Paneth cells (Figure 3).
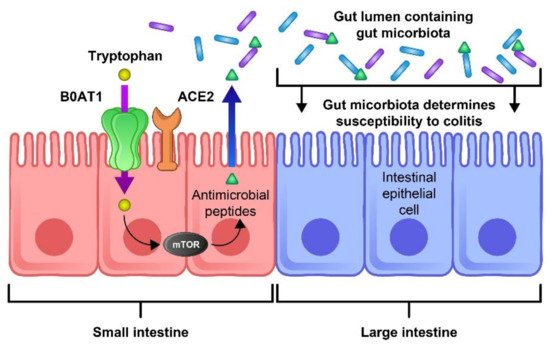
Figure 3. The role of ACE2 and mTOR activity in the expression of antimicrobial peptides and the composition of the gut flora. The ACE-2 receptor is essential for the surface expression of the neutral amino acid transporter B0AT1 in the small intestine. Tryptophan is mostly absorbed through the B0AT1/ACE2 pathway and activates the mTOR pathway, which regulates the expression of antimicrobial peptides. These peptides are important for maintaining an ideal microbiota in the large intestine. Thus, blocking this pathway by the binding of SARS-CoV-2 to the ACE-2 receptor could lead to inflammation, microbiota changes, and an increase in COVID-19 severity.
A recent study on the murine gut also reported that some microorganisms present in the intestine, such as Bacteroidesthetaiotaomicron, Bacteroidesmassiliensis, and Bacteroidesdorei, can downregulate ACE2 expression and inversely affect the SARS-CoV-2 load in a patient’s fecal samples [36].
This entry is adapted from the peer-reviewed paper 10.3390/jcm10214802
References
- Diao, K.; Han, P.; Pang, T.; Li, Y.; Yang, Z. HRCT imaging features in representative imported cases of 2019 novel coronavirus pneumonia. Precis. Clin. Med. 2020, 3, 9–13.
- Available online: https://www.who.int/data#reports (accessed on 23 August 2021).
- Moradian, N.; Ochs, H.D.; Sedikies, C.; Hamblin, M.R.; Camargo, C.A.; Martinez, J.A.; Biamonte, J.D.; Abdollahi, M.; Torres, P.J.; Nieto, J.J.; et al. The urgent need for integrated science to fight COVID-19 pandemic and beyond. J. Transl. Med. 2020, 18, 205.
- Hasan, A.; Paray, B.A.; Hussain, A.; Qadir, F.A.; Attar, F.; Aziz, F.M.; Sharifi, M.; Derakhshankhah, H.; Rasti, B.; Mehrabi, M.; et al. A review on the cleavage priming of the spike protein on coronavirus by angiotensin-converting enzyme-2 and furin. J. Biomol. Struct. Dyn. 2021, 39, 3025–3033.
- Tazikeh-Lemeski, E.; Moradi, S.; Raoufi, R.; Shahlaei, M.; Janlou, M.A.M.; Zolghadri, S. Targeting SARS-COV-2 non-structural protein 16: A virtual drug repurposing study. J. Biomol. Struct. Dyn. 2020, 1–14.
- Zhong, P.; Xu, J.; Yang, D.; Shen, Y.; Wang, L.; Feng, Y.; Du, C.; Song, Y.; Wu, C.; Hu, X.; et al. COVID-19-associated gastrointestinal and liver injury: Clinical features and potential mechanisms. Signal Transduct. Target. Ther. 2020, 5, 256.
- Bostanciklioglu, M. Temporal Correlation Between Neurological and Gastrointestinal Symptoms of SARS-CoV-2. Inflamm. Bowel Dis. 2020, 26, e89–e91.
- Jiehao, C.; Jin, X.; Daojiong, L.; Zhi, Y.; Lei, X.; Zhenghai, Q.; Yuehua, Z.; Hua, Z.; Ran, J.; Pengcheng, L.; et al. A Case Series of Children With 2019 Novel Coronavirus Infection: Clinical and Epidemiological Features. Clin. Infect. Dis. 2020, 71, 1547–1551.
- Wan, Y.; Li, J.; Shen, L.; Zou, Y.; Hou, L.; Zhu, L.; Faden, H.S.; Tang, Z.; Shi, M.; Jiao, N.; et al. Enteric involvement in hospitalised patients with COVID-19 outside Wuhan. Lancet 2020, 5, 534–535.
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus–Infected Pneumonia in Wuhan, China. JAMA 2020, 323, 1061–1069.
- Gou, W.; Fu, Y.; Yue, L.; Chen, G.-d.; Cai, X.; Shuai, M.; Xu, F.; Yi, X.; Chen, H.; Zhu, Y.; et al. Gut microbiota may underlie the predisposition of healthy individuals to COVID-19. medRxiv 2020.
- Marasco, G.; Lenti, M.V.; Cremon, C.; Barbaro, M.R.; Stanghellini, V.; Di Sabatino, A.; Barbara, G. Implications of SARS-CoV-2 infection for neurogastroenterology. Neurogastroenterol. Motil. 2021, 33, e14104.
- Chu, C.M.; Cheng, V.C.C.; Hung, I.F.N.; Wong, M.M.L.; Chan, K.H.; Chan, K.S.; Kao, R.Y.T.; Poon, L.L.M.; Wong, C.L.P.; Guan, Y.; et al. Role of lopinavir/ritonavir in the treatment of SARS: Initial virological and clinical findings. Thorax 2004, 59, 252–256.
- Young, B.E.; Ong, S.W.X.; Kalimuddin, S.; Low, J.G.; Tan, S.Y.; Loh, J.; Ng, O.-T.; Marimuthu, K.; Ang, L.W.; Mak, T.M.; et al. Epidemiologic Features and Clinical Course of Patients Infected With SARS-CoV-2 in Singapore. JAMA 2020, 323, 1488–1494.
- Wang, X.; Yang, C.; Sun, Y.; Sui, X.; Zhu, T.; Wang, Q.; Wang, S.; Yang, J.; Yang, W.; Liu, F.; et al. A novel screening strategy of anti-SARS-CoV-2 drugs via blocking interaction between Spike RBD and ACE2. Environ. Int. 2020, 147, 106361.
- Marjot, T.; Webb, G.J.; Barritt, A.S.; Moon, A.M.; Stamataki, Z.; Wong, V.W.; Barnes, E. COVID-19 and liver disease: Mechanistic and clinical perspectives. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 348–364.
- Chu, D.K.W.; Pan, Y.; Cheng, S.M.S.; Hui, K.P.Y.; Krishnan, P.; Liu, Y.; Ng, D.Y.M.; Wan, C.K.C.; Yang, P.; Wang, Q.; et al. Molecular Diagnosis of a Novel Coronavirus (2019-nCoV) Causing an Outbreak of Pneumonia. Clin. Chem. 2020, 66, 549–555.
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273.
- Li, W.; Moore, M.J.; Vasilieva, N.; Sui, J.; Wong, S.K.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greenough, T.C.; et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003, 426, 450–454.
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269.
- Galanopoulos, M.; Doukatas, A.; Gazouli, M. Origin and genomic characteristics of SARS-CoV-2 and its interaction with angiotensin converting enzyme type 2 receptors, focusing on the gastrointestinal tract. World J. Gastroenterol. 2020, 26, 6335–6345.
- Whittaker, G.R.; Daniel, S.; Millet, J.K. Coronavirus entry: How we arrived at SARS-CoV-2. Curr. Opin. Virol. 2021, 47, 113–120.
- Yuan, H.-W.; Wen, H.-L. Research progress on coronavirus S proteins and their receptors. Arch. Virol. 2021, 1–7.
- Yesudhas, D.; Srivastava, A.; Sekijima, M.; Gromiha, M.M. Tackling Covid-19 using disordered-to-order transition of residues in the spike protein upon angiotensin-converting enzyme 2 binding. Proteins 2021.
- Moradian, N.; Moallemian, M.; Delavari, F.; Sedikides, C.; Camargo, C.A.; Torres, P.J.; Sorooshian, A.; Mehdiabadi, S.P.; Nieto, J.J.; Bordas, S.; et al. Interdisciplinary Approaches to COVID-19. In Coronavirus Disease—COVID-19; Rezaei, N., Ed.; Springer International Publishing: Cham, Germany, 2021; pp. 923–936.
- Peron, J.P.S.; Nakaya, H. Susceptibility of the Elderly to SARS-CoV-2 Infection: ACE-2 Overexpression, Shedding, and Antibody-dependent Enhancement (ADE). Clinics 2020, 75, e1912.
- Zhang, H.; Kang, Z.; Gong, H.; Xu, D.; Wang, J.; Li, Z.; Li, Z.; Cui, X.; Xiao, J.; Zhan, J.; et al. Digestive system is a potential route of COVID-19: An analysis of single-cell coexpression pattern of key proteins in viral entry process. Gut 2020, 69, 1010.
- Xu, J.; Chu, M.; Zhong, F.; Tan, X.; Tang, G.; Mai, J.; Lai, N.; Guan, C.; Liang, Y.; Liao, G. Digestive symptoms of COVID-19 and expression of ACE2 in digestive tract organs. Cell Death Discov. 2020, 6, 76.
- Shafiee, S.; Cegolon, L.; Khafaei, M.; Gholami, N.; Zhao, S.; Khalesi, N.; Moosavian, H.; Fathi, S.; Izadi, M.; Ghadian, A.; et al. Gastrointestinal cancers, ACE-2/TMPRSS2 expression and susceptibility to COVID-19. Cancer Cell Int. 2021, 21, 431.
- Perisetti, A.; Goyal, H.; Gajendran, M.; Boregowda, U.; Mann, R.; Sharma, N. Prevalence, Mechanisms, and Implications of Gastrointestinal Symptoms in COVID-19. Front. Med. 2020, 7, 588711.
- Penninger, J.M.; Grant, M.B.; Sung, J.J.Y. The Role of Angiotensin Converting Enzyme 2 in Modulating Gut Microbiota, Intestinal Inflammation, and Coronavirus Infection. Gastroenterology 2021, 160, 39–46.
- Martinez-Guryn, K.; Leone, V.; Chang, E.B. Regional diversity of the gastrointestinal microbiome. Cell Host Microbe 2019, 26, 314–324.
- He, L.H.; Ren, L.F.; Li, J.F.; Wu, Y.N.; Li, X.; Zhang, L. Intestinal Flora as a Potential Strategy to Fight SARS-CoV-2 Infection. Front. Microbiol. 2020, 11, 1388.
- Villapol, S. Gastrointestinal symptoms associated with COVID-19: Impact on the gut microbiome. Transl. Res. 2020, 226, 57–69.
- Rajput, S.; Paliwal, D.; Naithani, M.; Kothari, A.; Meena, K.; Rana, S. COVID-19 and Gut Microbiota: A Potential Connection. Indian J. Clin. Biochem. 2021, 36, 1–12.
- Zuo, T.; Zhang, F.; Lui, G.C.Y.; Yeoh, Y.K.; Li, A.Y.L.; Zhan, H.; Wan, Y.; Chung, A.C.K.; Cheung, C.P.; Chen, N.; et al. Alterations in Gut Microbiota of Patients With COVID-19 During Time of Hospitalization. Gastroenterology 2020, 159, 944–955.e948.
- Sandhu, A.; Tillotson, G.; Polistico, J.; Salimnia, H.; Cranis, M.; Moshos, J.; Cullen, L.; Jabbo, L.; Diebel, L.; Chopra, T. Clostridioides difficile in COVID-19 Patients, Detroit, Michigan, USA, March–April 2020. Emerg. Infect. Dis. 2020, 26.
- Dhar, D.; Mohanty, A. Gut microbiota and Covid-19- possible link and implications. Virus Res. 2020, 285, 198018.
- Syed, A.; Khan, A.; Gosai, F.; Asif, A.; Dhillon, S. Gastrointestinal pathophysiology of SARS-CoV2—A literature review. J. Community Hosp. Intern. Med. Perspect 2020, 10, 523–528.
- Kawai, T.; Akira, S. The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat. Immunol. 2010, 11, 373.
- Pawlik, M.W.; Kwiecien, S.; Ptak-Belowska, A.; Pajdo, R.; Olszanecki, R.; Suski, M.; Madej, J.; Targosz, A.; Konturek, S.J.; Korbut, R. The renin-angiotensin system and its vasoactive metabolite angiotensin-(1-7) in the mechanism of the healing of preexisting gastric ulcers. The involvement of Mas receptors, nitric oxide, prostaglandins and proinflammatory cytokines. J. Physiol. Pharm. 2016, 67, 75–91.
- Fujita, M.; Hayashi, I.; Yamashina, S.; Fukamizu, A.; Itoman, M.; Majima, M. Angiotensin type 1a receptor signaling-dependent induction of vascular endothelial growth factor in stroma is relevant to tumor-associated angiogenesis and tumor growth. Carcinogenesis 2005, 26, 271–279.
- Rahban, M.; Habibi-Rezaei, M.; Mazaheri, M.; Saso, L.; Moosavi-Movahedi, A.A. Anti-Viral. Potential and Modulation of Nrf2 by Curcumin: Pharmacological Implications. Antioxidants 2020, 9, 1228.
- Raimondo, M.G.; Biggioggero, M.; Crotti, C.; Becciolini, A.; Favalli, E.G. Profile of sarilumab and its potential in the treatment of rheumatoid arthritis. Drug Des. Dev. 2017, 11, 1593–1603.
- Lowery, S.A.; Sariol, A.; Perlman, S. Innate immune and inflammatory responses to SARS-CoV-2: Implications for COVID-19. Cell Host Microbe 2021, 29, 1052–1062.
- Garg, M.; Royce, S.; Tikellis, C.; Shallue, C.; Batu, D.; Velkoska, E.; Burrell, L.; Patel, S.; Beswick, L.; Jackson, A.; et al. Imbalance of the renin-angiotensin system may contribute to inflammation and fibrosis in IBD: A novel therapeutic target? Gut 2019, 69, 841–851.
- Liang, W.; Feng, Z.; Rao, S.; Xiao, C.; Xue, X.; Lin, Z.; Zhang, Q.; Qi, W. Diarrhoea may be underestimated: A missing link in 2019 novel coronavirus. Gut 2020, 69, 1141.
- D’Amico, F.; Baumgart, D.C.; Danese, S.; Peyrin-Biroulet, L. Diarrhea during COVID-19 infection: Pathogenesis, epidemiology, prevention and management. Clin. Gastroenterol. Hepatol. 2020, 18, 1663–1672.
- Tang, W.; Cao, Z.; Han, M.; Wang, Z.; Chen, J.; Sun, W.; Wu, Y.; Xiao, W.; Liu, S.; Chen, E.; et al. Hydroxychloroquine in patients with mainly mild to moderate coronavirus disease 2019: Open label, randomised controlled trial. BMJ 2020, 369, m1849.
- Chedid, M.; Waked, R.; Haddad, E.; Chetata, N.; Saliba, G.; Choucair, J. Antibiotics in treatment of COVID-19 complications: A review of frequency, indications, and efficacy. J. Infect. Public Health 2021, 14, 570.
- Bagheri, A.; Moezzi, S.M.I.; Mosaddeghi, P.; Parashkouhi, S.N.; Hoseini, S.M.F.; Badakhshan, F.; Negahdaripour, M. Interferon-inducer antivirals: Potential candidates to combat COVID-19. Int. Immunopharmacol. 2021, 91, 107245.
- Frediansyah, A.; Tiwari, R.; Sharun, K.; Dhama, K.; Harapan, H. Antivirals for COVID-19: A critical review. Clin. Epidemiol. Glob. Health 2021, 9, 90–98.
- Belkaid, Y.; Harrison, O.J. Homeostatic Immunity and the Microbiota. Immunity 2017, 46, 562–576.
- Selber-Hnatiw, S.; Rukundo, B.; Ahmadi, M.; Akoubi, H.; Al-Bizri, H.; Aliu, A.F.; Ambeaghen, T.U.; Avetisyan, L.; Bahar, I.; Baird, A.; et al. Human Gut Microbiota: Toward an Ecology of Disease. Front. Microbiol. 2017, 8, 1265.
- Gong, S.; Lan, T.; Zeng, L.; Luo, H.; Yang, X.; Li, N.; Chen, X.; Liu, Z.; Li, R.; Win, S.; et al. Gut microbiota mediates diurnal variation of acetaminophen induced acute liver injury in mice. J. Hepatol. 2018, 69, 51–59.
- Abt, M.C.; Osborne, L.C.; Monticelli, L.A.; Doering, T.A.; Alenghat, T.; Sonnenberg, G.F.; Paley, M.A.; Antenus, M.; Williams, K.L.; Erikson, J.; et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity 2012, 37, 158–170.
- Abrahamsson, T.R.; Jakobsson, H.E.; Andersson, A.F.; Björkstén, B.; Engstrand, L.; Jenmalm, M.C. Low gut microbiota diversity in early infancy precedes asthma at school age. Clin. Exp. Allergy 2014, 44, 842–850.
- Donaldson, G.P.; Lee, S.M.; Mazmanian, S.K. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 2016, 14, 20–32.
- Dickson, R.P.; Singer, B.H.; Newstead, M.W.; Falkowski, N.R.; Erb-Downward, J.R.; Standiford, T.J.; Huffnagle, G.B. Enrichment of the lung microbiome with gut bacteria in sepsis and the acute respiratory distress syndrome. Nat. Microbiol. 2016, 1, 16113.
- Kim, H.S. Do an altered gut microbiota and an associated leaky gut affect COVID-19 severity? Mbio 2021, 12, e03022-20.
- Li, J.; Richards, E.M.; Handberg, E.M.; Pepine, C.J.; Raizada, M.K. Butyrate regulates COVID-19–relevant genes in gut epithelial organoids from normotensive rats. Hypertension 2021, 77, e13–e16.
- Bradley, C.P.; Teng, F.; Felix, K.M.; Sano, T.; Naskar, D.; Block, K.E.; Huang, H.; Knox, K.S.; Littman, D.R.; Wu, H.J. Segmented Filamentous Bacteria Provoke Lung Autoimmunity by Inducing Gut-Lung Axis Th17 Cells Expressing Dual TCRs. Cell Host Microbe 2017, 22, 697–704.e694.
- Ye, L.; Yang, Z.; Liu, J.; Liao, L.; Wang, F. Digestive system manifestations and clinical significance of coronavirus disease 2019: A systematic literature review. J. Gastroenterol. Hepatol. 2021, 36, 1414–1422.
- Liu, S.; Tang, M.-M.; Du, J.; Gong, Z.-C.; Sun, S.-S. COVID-19 in gastroenterology and hepatology: Lessons learned and questions to be answered. World J. Clin. Cases 2021, 9, 4199.
- Hashimoto, T.; Perlot, T.; Rehman, A.; Trichereau, J.; Ishiguro, H.; Paolino, M.; Sigl, V.; Hanada, T.; Hanada, R.; Lipinski, S.; et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature 2012, 487, 477–481.
- Manosso, L.M.; Arent, C.O.; Borba, L.A.; Ceretta, L.B.; Quevedo, J.; Réus, G.Z. Microbiota-Gut-Brain Communication in the SARS-CoV-2 Infection. Cells 2021, 10, 1993.
This entry is offline, you can click here to edit this entry!
