The application of aptamers in biomedicine is emerging as an essential technology in the field of cancer research. As small single-stranded DNA or RNA ligands with high specificity and low immunogenicity for their targets, aptamers provide many advantages in cancer therapeutics over protein-based molecules, such as antibodies. Vimentin is an intermediate filament protein that is overexpressed in endothelial cells of cancerous tissue. High expression levels of vimentin have been associated with increased capacity for migration and invasion of the tumor cells. We have selected and identified thioated aptamers with high specificity for vimentin using human ovarian cancer tissues. Tentative binding motifs were chosen for two vimentin aptamers based on predicted secondary structures. Each of these shorter, tentative binding motifs was synthesized, purified, and characterized via cell binding assays. Two vimentin binding motifs with high fidelity binding were selected and further characterized via cell and tissue binding assays, as well as flow cytometric analysis. The equilibrium binding constants of these small thioated aptamer constructs were also determined. Future applications for the vimentin binding aptamer motifs include conjugation of the aptamers to synthetic dyes for use in targeted imaging and therapy, and ultimately more detailed and precise monitoring of treatment response and tumor progression in ovarian pathology.
- aptamer
- binding motifs
- ovarian cancer
[1][2][3][4][5][6][7][8][9][10][11][12][13][14][15][16][17][18][19][20][21][22][23][24][25]
1. Introduction
2. Results
2.1. Identification of Potential Aptamer Binding Motifs
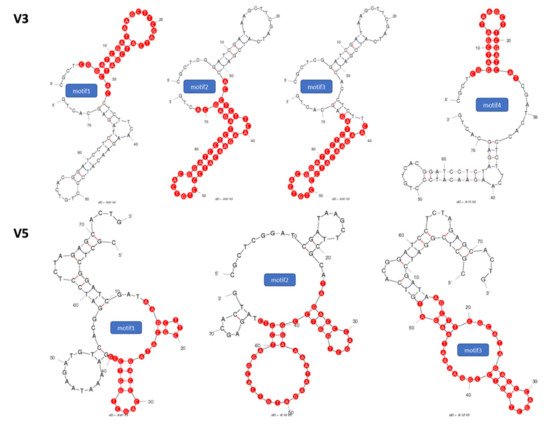
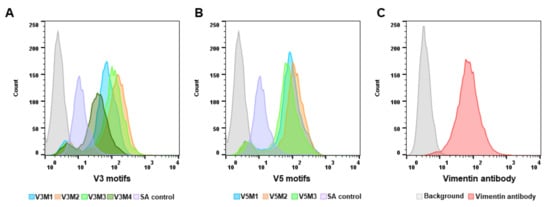
2.2. Screening of Synthesized Aptamer Motifs
| Name | Sequences |
| V3M2 (40 mer) | 5'-ACCTCTTCAAGAACATCCCTGTCACGGATCCTCTAGAGCA-3' |
| V5M2 (41 mer) | 5'-TAGACCCAGCTGGTCCGGAAAATAAGATGTCACGGATCCTC-3' |
| Scrambled Control (40 mer) | 5'-CCCACTTATCGTCCCTTAATGAGTTTACTCGCACACCGGA-3' |
2.3. Binding Affinity of Selected V3M2 and V5M2
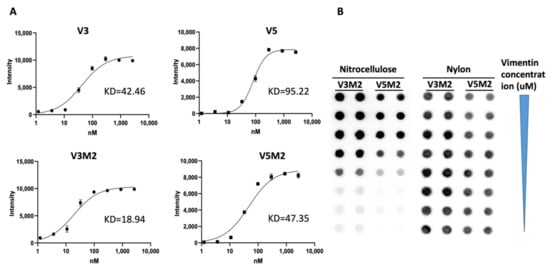
2.4. Dose Response of V3M2 and V5M2
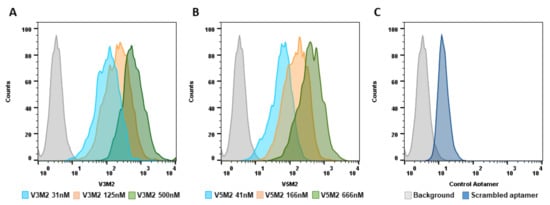
2.5. Validation of Selected Motifs with Human Cells
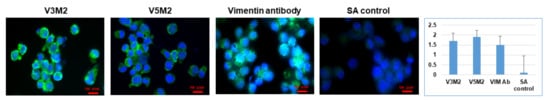
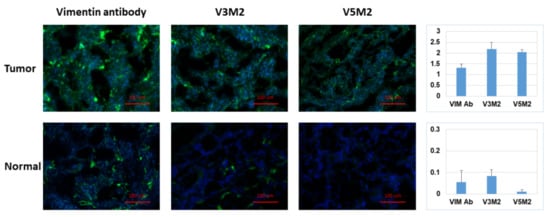
2.6. Validation of Selected Motifs with Human Ovarian Tumor
3. Current Insights
This entry is adapted from the peer-reviewed paper 10.3390/molecules26216525
References
- [5] M. Zuker, “Mfold web server for nucleic acid folding and hybridization prediction,” Nucleic Acids Research, vol. 31, no. 13, pp. 3406-3415, 2003.
- [6] P. Dao, J. Hoinka, M. Takahashi, J. Zhou, M. Ho, Y. Wang, F. Costa, J. J. Rossi, R. Backofen, J. Burnett, and T. M. Przytycka, “AptaTRACE Elucidates RNA Sequence-Structure Motifs from Selection Trends in HT-SELEX Experiments,” Cell Syst, vol. 3, no. 1, pp. 62-70, Jul, 2016.
- [7] J. Hoinka, E. Zotenko, A. Friedman, Z. E. Sauna, and T. M. Przytycka, “Identification of sequence-structure RNA binding motifs for SELEX-derived aptamers,” Bioinformatics, vol. 28, no. 12, pp. i215-23, Jun 15, 2012.
- [8] J. Caroli, C. Taccioli, A. De La Fuente, P. Serafini, and S. Bicciato, “APTANI: a computational tool to select aptamers through sequence-structure motif analysis of HT-SELEX data,” Bioinformatics, vol. 32, no. 2, pp. 161-164, 2015.
- [9] T. Adachi, and Y. Nakamura, “Aptamers: A Review of Their Chemical Properties and Modifications for Therapeutic Application,” Molecules (Basel, Switzerland), vol. 24, no. 23, pp. 4229, 2019.
- [10] S. P. Henry, P. C. Giclas, J. Leeds, M. Pangburn, C. Auletta, A. A. Levin, and D. J. Kornbrust, “Activation of the alternative pathway of complement by a phosphorothioate oligonucleotide: potential mechanism of action,” J Pharmacol Exp Ther, vol. 281, no. 2, pp. 810-6, May, 1997.
- [11] C. Qi, T. Bing, H. Mei, X. Yang, X. Liu, and D. Shangguan, “G-quadruplex DNA aptamers for zeatin recognizing,” Biosens Bioelectron, vol. 41, pp. 157-62, Mar 15, 2013.
- [12] H. Mei, T. Bing, X. Yang, C. Qi, T. Chang, X. Liu, Z. Cao, and D. Shangguan, “Functional-group specific aptamers indirectly recognizing compounds with alkyl amino group,” Anal Chem, vol. 84, no. 17, pp. 7323-9, Sep 4, 2012.
- [13] H. Kaur, and L. Y. Yung, “Probing high affinity sequences of DNA aptamer against VEGF165,” PLoS One, vol. 7, no. 2, pp. e31196, 2012.
- [14] G. V. Kupakuwana, J. E. Crill, 2nd, M. P. McPike, and P. N. Borer, “Acyclic identification of aptamers for human alpha-thrombin using over-represented libraries and deep sequencing,” PLoS One, vol. 6, no. 5, pp. e19395, 2011.
- [15] W. H. Thiel, T. Bair, K. Wyatt Thiel, J. P. Dassie, W. M. Rockey, C. A. Howell, X. Y. Liu, A. J. Dupuy, L. Huang, R. Owczarzy, M. A. Behlke, J. O. McNamara, and P. H. Giangrande, “Nucleotide bias observed with a short SELEX RNA aptamer library,” Nucleic Acid Ther, vol. 21, no. 4, pp. 253-63, Aug, 2011.
- [16] J. Macdonald, P. Houghton, D. Xiang, W. Duan, and S. Shigdar, “Truncation and Mutation of a Transferrin Receptor Aptamer Enhances Binding Affinity,” Nucleic Acid Ther, vol. 26, no. 6, pp. 348-354, Dec, 2016.
- [17] J. E. Eriksson, T. Dechat, B. Grin, B. Helfand, M. Mendez, H.-M. Pallari, and R. D. Goldman, “Introducing intermediate filaments: from discovery to disease,” The Journal of clinical investigation, vol. 119, no. 7, pp. 1763-1771, 2009.
- [18] L. Zhao, P. Zhang, X.-j. Su, and B. Zhang, “The ubiquitin ligase TRIM56 inhibits ovarian cancer progression by targeting vimentin,” Journal of Cellular Physiology, vol. 233, no. 3, pp. 2420-2425, 2018.
- [19] L. Du, J. Li, L. Lei, H. He, E. Chen, J. Dong, and J. Yang, “High Vimentin Expression Predicts a Poor Prognosis and Progression in Colorectal Cancer: A Study with Meta-Analysis and TCGA Database,” BioMed research international, vol. 2018, pp. 6387810-6387810, 2018.
- [20] S. Yin, F. F. Chen, and G. F. Yang, “Vimentin immunohistochemical expression as a prognostic factor in gastric cancer: A meta-analysis,” Pathol Res Pract, vol. 214, no. 9, pp. 1376-1380, Sep, 2018.
- [21] S. H. Zou S, Fan L, Xiao X, Gong L, Zhu J, Chen X, “Prognostic indicators in patients with early stage endometrioid adenocarcinoma: a retrospective case-control study of 523 patients,” Int J Clin Exp Med, vol. 10, no. 2, pp. 3699-3705, 2017.
- [22] S. Szubert, K. Koper, M. M. Dutsch-Wicherek, and W. Jozwicki, “High tumor cell vimentin expression indicates prolonged survival in patients with ovarian malignant tumors,” Ginekologia Polska, vol. 90, no. 1, pp. 11-19, 2019-01-31, 2019.
- [23] Y. Zheng, J. Zhang, M. Huang, T. Wang, X. Qu, L. Wu, J. Song, W. Wang, Y. Song, and C. Yang, “Selection of Aptamers Against Vimentin for Isolation and Release of Circulating Tumor Cells Undergoing Epithelial Mesenchymal Transition,” Anal Chem, vol. 92, no. 7, pp. 5178-5184, Apr 7, 2020.
- [24] T. N. Zamay, O. S. Kolovskaya, Y. E. Glazyrin, G. S. Zamay, S. A. Kuznetsova, E. A. Spivak, M. Wehbe, A. G. Savitskaya, O. A. Zubkova, A. Kadkina, X. Wang, D. Muharemagic, A. Dubynina, Y. Sheina, A. B. Salmina, M. V. Berezovski, and A. S. Zamay, “DNA-aptamer targeting vimentin for tumor therapy in vivo,” Nucleic acid therapeutics, vol. 24, no. 2, pp. 160-170, 2014.
- [25] H. Wang, X. Li, D. E. Volk, G. L. Lokesh, M. A. Elizondo-Riojas, L. Li, A. M. Nick, A. K. Sood, K. P. Rosenblatt, and D. G. Gorenstein, “Morph-X-Select: Morphology-based tissue aptamer selection for ovarian cancer biomarker discovery,” Biotechniques, vol. 61, no. 5, pp. 249-259, 2016.
- [26] D. Coppola, L. Fu, S. V. Nicosia, S. Kounelis, and M. Jones, “Prognostic significance of p53, bcl-2, vimentin, and S100 protein-positive Langerhans cells in endometrial carcinoma,” Hum Pathol, vol. 29, no. 5, pp. 455-62, May, 1998.
- [27] A. Satelli, and S. Li, “Vimentin in cancer and its potential as a molecular target for cancer therapy,” Cellular and molecular life sciences : CMLS, vol. 68, no. 18, pp. 3033-3046, 2011.
- [28] H. J. Gaus, R. Gupta, A. E. Chappell, M. E. Østergaard, E. E. Swayze, and P. P. Seth, “Characterization of the interactions of chemically-modified therapeutic nucleic acids with plasma proteins using a fluorescence polarization assay,” Nucleic Acids Res, vol. 47, no. 3, pp. 1110-1122, Feb 20, 2019.
- [29] R. E. Armstrong, and G. F. Strouse, “Rationally manipulating aptamer binding affinities in a stem-loop molecular beacon,” Bioconjugate chemistry, vol. 25, no. 10, pp. 1769-1776, 2014.
- [26] D. Coppola, L. Fu, S. V. Nicosia, S. Kounelis, and M. Jones, “Prognostic significance of p53, bcl-2, vimentin, and S100 protein-positive Langerhans cells in endometrial carcinoma,” Hum Pathol, vol. 29, no. 5, pp. 455-62, May, 1998.
- [27] A. Satelli, and S. Li, “Vimentin in cancer and its potential as a molecular target for cancer therapy,” Cellular and molecular life sciences : CMLS, vol. 68, no. 18, pp. 3033-3046, 2011.
- [28] H. J. Gaus, R. Gupta, A. E. Chappell, M. E. Østergaard, E. E. Swayze, and P. P. Seth, “Characterization of the interactions of chemically-modified therapeutic nucleic acids with plasma proteins using a fluorescence polarization assay,” Nucleic Acids Res, vol. 47, no. 3, pp. 1110-1122, Feb 20, 2019.
- [29] R. E. Armstrong, and G. F. Strouse, “Rationally manipulating aptamer binding affinities in a stem-loop molecular beacon,” Bioconjugate chemistry, vol. 25, no. 10, pp. 1769-1776, 2014.
