Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Virology
Noroviruses (NoVs) are a group of non-enveloped RNA viruses belonging to the Norovirus genus in the Caliciviridae family, and they are the leading cause of sporadic and epidemic nonbacterial acute gastroenteritis (AGE) in humans.
- norovirus
- GII.3 genotype
- monoclonal antibody
1. Introduction
Noroviruses (NoVs) are a group of non-enveloped RNA viruses belonging to the Norovirus genus in the Caliciviridae family, and they are the leading cause of sporadic and epidemic nonbacterial acute gastroenteritis (AGE) in humans [1][2][3]. NoVs possess a single-stranded, positive-sense RNA genome about 7.5 ~ 7.7 kb in length [4][5], which contains three open reading frames (ORF): ORF1 encodes the replicase polyprotein, ORF2 encodes the major capsid protein named VP1, and ORF3 encodes the minor capsid protein named VP2 [4][6][7]. VP1 capsid protein consists of a shell (S) domain and a protruding (P) domain that can be further divided into two subdomains, namely P1 and P2 [4][6][7]. The P2 domain of most NoVs harbors binding sites for human histo-blood group antigens (HBGAs) [8][9][10], which are complex, fucose-containing carbohydrates present abundantly on the intestinal epithelia and function as an attachment receptor for human NoVs [11][12][13].
Based on the VP1 amino acid sequence, NoVs were classified into six genogroups (GI to GVI) in 2013 [14]. This NoV classification scheme was recently updated, with the number of genogroups expanded to 10 (GI to GX) [3][14]. Viruses of GI, GII, and GIV infect humans, and in particular, GII, which comprises 27 genotypes [14], accounts for approximately 90% of norovirus infections in humans [15]. Among all GII genotypes, GII.4 has been the predominant one causing AGE in humans of all ages over the past two decades [16][17][18][19][20], while GII.3 is one of the most common genotypes associated with NoV infection in infants and young children [21][22][23][24][25][26][27][28][29][30]. In particular, one clinical study showed that GII.3 and GII.4 were responsible for 71.24% and 23.53% of NoV-associated pediatric AGE, respectively, in Hohhot, China, between January 2012 and December 2017 [27]. It was estimated that 70% of children would have been infected by GII.3 by 2 years of age [31].
GII.3 NoVs undergo constant evolution, driven primarily by intergenic recombination [21][22][32]. The initial phylogenetic analysis, which was published in 2011, divided GII.3 NoVs into three clusters (I, II, and III) based on the available 63 GII.3 VP1 sequences [21]. Two years later, these relatively larger clusters were further defined into five smaller lineages (A to E), which were generally observed to be temporally sequential in terms of collection dates of the corresponding strains within each lineage [22]. In 2020, Saito et al. performed a phylogenetic analysis of a large number of sequences of GII.3 strains, most of which were collected after 2013, and therefore updated GII.3 classification with the analyzed strains being divided into three clusters (1, 2, and 3) based on the VP1 amino acid sequence [33].
The GII.3 VP1 protein can self-assemble into virus-like particles [34][35][36], with the outer P domain in either “resting” or “rising” conformation depending on the pH of sample solutions [35]. GII.3 VLPs formed by the entire VP1 protein or P particles solely made of the P domain can bind HBGAs in vitro [8][21][34][37]. A recent structural study has defined an HBGA binding site on GII.3, which is constituted by eight VP1 residues within the P2 subdomain [36]. Immunization of animals with GII.3 VLPs elicited antibodies capable of blocking the interaction between HBGAs and homotypic VLPs [34][37], indicating that the GII.3 VP1 protein does contain blockade antibody epitopes. Although in silico analyses have predicted several sites where GII.3 blockade antibody epitopes might reside [21][22][33], thus far, the exact locations of GII.3 blockade antibody epitopes have not been defined experimentally.
2. Generation and Characterization of the Anti-GII.3 mAbs
To develop GII.3-specific mAbs, we immunized Balb/c mice with VLP of the GII.3 cluster 3 strain Hu/GII.3/CUHK-NS-227/2014/CHN (GenBank ID: KJ499444; designated as GII.3-KJ, Table 1). Splenocytes from the VLP-immunized mice were fused with SP2/0 myeloma cells to generate hybridomas. Culture supernatants from the resulting hybridomas were analyzed by ELISA for their reactivity with the immunogen VLP. The results showed that four individual hybridoma clones, designated 3A3, 8C7, 8D1, and 9B8, were ELISA-positive (Table 2). Isotyping analysis revealed that all of these four clones were IgG1 antibodies (Table 2).
Table 1. A summary of all the GII.3 VLPs used in this study.
| VLP | Strain | GenBank ID | Cluster b |
|---|---|---|---|
| GII.3-KJ a | Hu/GII.3/CUHK-NS-227/2014/CHN | KJ499444 | 3 |
| GII.3-KY | Hu/GII.3/3-34/2015/HNZZ/CHN | KY767665 | 2 |
| GII.3-AB38 | Hu/GII.3/RotterdamP8D31/2006/NL | AB385642 | 2 |
| GII.3-AB06 | Hu/NoV/GII.3/SaitamaU201/1998/JPN | AB067542 | 1 |
a GII.3-KJ VLP was used to produce the mAbs. b According to Saito et al. [33].
Table 2. Characteristics of the anti-GII.3 mAbs.
| MAb | Isotype | Binding Activity to GII.3 VLP a | Binding Affinity with GII.3 VLP b | ||
|---|---|---|---|---|---|
| KD (nM) | Kon (1/Ms) | Kdis (1/s) | |||
| 8C7 | IgG1 | + | <0.001 | 1.88 × 105 | <1.0 × 10−7 |
| 8D1 | IgG1 | + | <0.001 | 3.14 × 105 | <1.0 × 10−7 |
| 9B8 | IgG1 | + | 0.161 | 4.15 × 105 | 6.66 × 10−5 |
| 3A3 | IgG1 | + | 31.0 | 6.1 × 103 | 1.89 × 10−4 |
a Hybridoma culture supernatants were tested for binding to GII.3 VLP by ELISA. +, OD450 > 0.5. b Binding affinities of the MAbs to GII.3 VLP were measured by BLI. Related to Figure 1.
We performed biolayer interferometry (BLI) assays to determine the binding affinity of the mAbs to the GII.3 VLP. The results showed that 8C7 and 8D1 possessed high affinity to the GII.3 VLP with KD values of <0.001 nM, whereas 3A3 and 9B8 had a much lower affinity with KD values being 31 and 0.161 nM, respectively (Table 2). Representative BLI graphs for 8C7 and 8D1 mAbs are shown in Figure 1A,B.
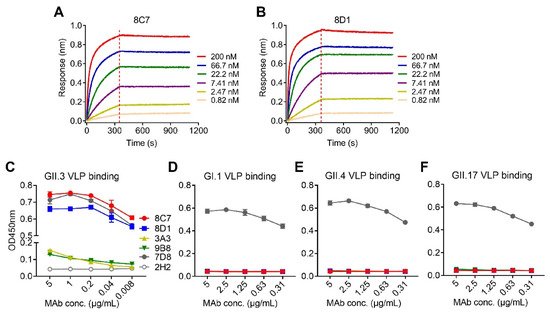
Figure 1. Binding affinity and specificity of the mAbs to GII.3 VLP. (A,B) Biolayer interferometry sensorgrams of anti-GII.3 mAbs 8C7 (A) and 8D1 (B) binding to biotinylated GII.3-KJ VLP on streptavidin-coated biosensors. Association and dissociation steps are divided by dashed lines. The mAb concentrations were shown. (C–F) Reactivity of anti-GII.3 mAbs (8C7, 8D1, 3A3, and 9B8) with GII.3-KJ VLP (C), GI.1 VLP (D), GII.4 VLP (E), and GII.17 VLP (F) were determined by ELISA. Data are mean ± SD of triplicate wells. 7D8, a norovirus cross-reactive antibody, used as a positive control. 2H2, an anti-SARS-CoV-2 mAb, used as a negative control. Conc., the abbreviation for concentration.
Then, authors assessed the binding specificity of the four mAbs by performing ELISAs with VLPs of four different genotypes (GII.3, GII.4, GII.17, and GI.1) as coating antigen (Figure 1C–F). MAbs 8C7 and 8D1 strongly reacted with the GII.3 VLP in an antibody dose-dependent manner, whereas only high concentrations of 3A3 and 9B8 yield detectable reactivity with the GII.3 VLP (Figure 1C), in consistency with their binding affinity measured by BLI (Table 2 and Figure 1A,B). In contrast, none of the four anti-GII.3 mAbs showed binding activity to the GI.1 VLP, the GII.4 VLP, or the GII.17 VLP, regardless of the antibody concentration (Figure 1D–F). As the positive control, a previously identified norovirus cross-reactive mAb, 7D8, was found to efficiently bind with each of the four VLPs (Figure 1C–F), validating the assays. Based on the above data, we conclude that the four mAbs generated here are indeed GII.3 specific.
3. Blockade Activity of the Four Anti-GII.3 mAbs
HBGAs are considered a binding receptor for NoVs [38][39]. Vaccine-elicited antibody titers that blocked VLPs binding with HBGAs correlated with protection in clinical trials [40][41], and hence HBGA blockade ELISAs have been used as a surrogate neutralization assay to assess the efficacy of candidate vaccines and antibodies [42][43]. To evaluate the functions of the four anti-GII.3 mAbs, we performed blockade ELISAs with pig gastric mucin (PGM) type III as the source of HBGAs [44]. As shown in Figure 2, 8C7 and 8D1 mAbs efficiently blocked the immunogen GII.3 VLP binding to PGM with IC50s of 0.24 µg/mL and 0.19 µg/mL, respectively, whereas no blocking effect was observed for 3A3 and 9B8 as well as for the isotype control mAb, regardless of the antibody concentrations used. These data demonstrate that 8C7 and 8D1, but not 3A3 and 9B8, are potent blockade mAbs. Hence, we focused our subsequent analyses on 8C7 and 8D1.
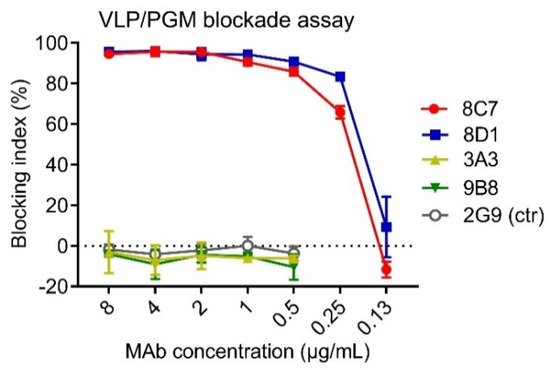
Figure 2. The ability of the mAbs to block the interaction between GII.3-KJ VLP and HBGA-containing PGM III was tested by ELISA. HBsAg-specific mAb 2G9 was used as a control (ctr). Blocking index (%) was determined by comparing the results of the VLP/MAb wells with the VLP-only wells. Data are mean ± SD of triplicate wells.
4. Intratypic Cross-Binding by 8C7 and 8D1 mAbs
To examine the mAbs’ cross-reactivity within GII.3 genotype, VLPs of the strains GII.3-KY (cluster 2, according to Saito et al. [33]), GII.3-AB38 (cluster 2), and GII.3-AB06 (cluster 1) were produced (Table 1). These VLPs were compared with the immunogen VLP (GII.3-KJ, cluster 3) by ELISA for their reactivity with 8C7, 8D1, or the rabbit anti-GII.3-KJ VLP sera. As expected, the rabbit anti-GII.3 sera could react with all VLPs, serving as a positive control in this assay (Figure 3). The 8C7 and 8D1 mAbs efficiently bound the original (GII.3-KJ) VLP and the GII.3-KY VLP but did not show significant reactivity to the VLPs of GII.3-AB38 and GII.3-AB06 (Figure 3).
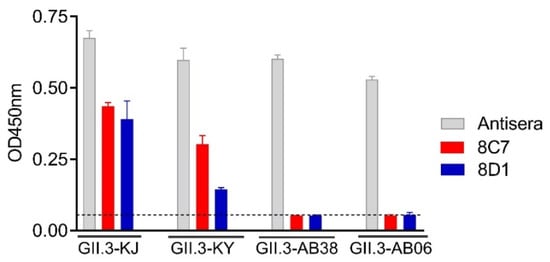
Figure 3. Cross-reactivity of the 8C7 and 8D1 mAbs was tested by ELISA with various wild-type GII.3 VLPs. Rabbit anti-GII.3 VLP antiserum was used as a positive control. Dashed line indicates the value from the blank well (containing no antigen). Data are mean ± SD of triplicate wells.
5. Epitope Mapping for 8C7 and 8D1 mAbs
To roughly locate the epitopes of blockade antibodies 8C7 and 8D1, we used GII.3-AB38, which did not react with the two mAbs, as the backbone to construct a series of chimeric VLPs. Briefly, fragments of the GII.3-AB38 VP1 protein were replaced with the corresponding ones from the immunogen strain GII.3-KJ (Figure 4A). The resulting chimeric VLPs were analyzed for their reactivity with 8C7, 8D1, and the anti-GII.3 sera by ELISA. The results were summarized in Figure 4A, and representative data were shown in Figure 4B. Specifically, GII.3-KJ(268–420), which contained the exact P2 domain (amino acids 268–420) of the GII.3-KJ VP1 protein, could react with both 8C7 and 8D1, indicating that the P2 domain harbors the epitopes of the two mAbs. GII.3-KJ(351–548) and GII.3-KJ(384–548) positively reacted with 8C7 whereas GII.3-KJ(400–548) failed to do (Figure 4A,B), suggesting that the 8C7 epitope may be located within residues 384–400. Meanwhile, 8D1 mAb bound to each of the above four chimeric VLPs, inferring that the 8D1 epitope may encompass residues 401–420.
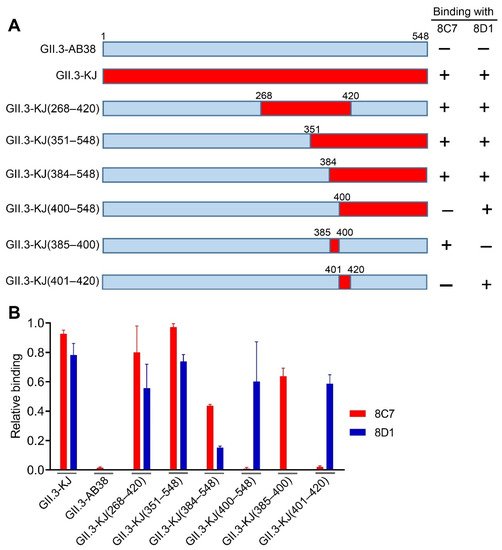
Figure 4. Epitope mapping of mAbs 8C7 and 8D1. (A) Diagram of the parental and chimeric GII.3 VLPs. The summary of the VLP-binding ELISA results is shown on the right-hand side. +, positive result; –, negative result. (B) Binding activities of the 8C7 and 8D1 mAbs to the indicated parental and chimeric GII.3 VLPs were tested by ELISA. For a given VLP sample, its reactivity with 8C7 or 8D1 was normalized against that with rabbit polyclonal antibody raised against GII.3 VLP, and relative binding was calculated as follows: (OD450 value obtained with mAb—OD450 value of blank well)/(OD450 value obtained with anti-GII.3 polyclonal antibody—OD450 value of blank well). Data are mean ± SEM of triplicate samples.
This entry is adapted from the peer-reviewed paper 10.3390/v13102058
References
- Lindesmith, L.; Moe, C.; Marionneau, S.; Ruvoen, N.; Jiang, X.; Lindblad, L.; Stewart, P.; Le Pendu, J.; Baric, R. Human susceptibility and resistance to Norwalk virus infection. Nat. Med. 2003, 9, 548–553.
- Green, K.Y. The role of human caliciviruses in epidemic gastroenteritis. Arch. Virol. Suppl. 1997, 13, 153–165.
- Chhabra, P.; de Graaf, M.; Parra, G.I.; Chan, M.C.; Green, K.; Martella, V.; Wang, Q.; White, P.A.; Katayama, K.; Vennema, H.; et al. Updated classification of norovirus genogroups and genotypes. J. Gen. Virol. 2019, 100, 1393–1406.
- Jiang, X.; Wang, M.; Wang, K.; Estes, M.K. Sequence and genomic organization of Norwalk virus. Virology 1993, 195, 51–61.
- Katayama, K.; Shirato-Horikoshi, H.; Kojima, S.; Kageyama, T.; Oka, T.; Hoshino, F.; Fukushi, S.; Shinohara, M.; Uchida, K.; Suzuki, Y.; et al. Phylogenetic analysis of the complete genome of 18 Norwalk-like viruses. Virology 2002, 299, 225–239.
- Hardy, M.E. Norovirus protein structure and function. FEMS Microbiol. Lett. 2005, 253, 1–8.
- Prasad, B.V.; Hardy, M.E.; Dokland, T.; Bella, J.; Rossmann, M.G.; Estes, M.K. X-ray crystallographic structure of the Norwalk virus capsid. Science 1999, 286, 287–290.
- Tan, M.; Jiang, X. The p domain of norovirus capsid protein forms a subviral particle that binds to histo-blood group antigen receptors. J. Virol. 2005, 79, 14017–14030.
- Cao, S.; Lou, Z.; Tan, M.; Chen, Y.; Liu, Y.; Zhang, Z.; Zhang, X.C.; Jiang, X.; Li, X.; Rao, Z. Structural basis for the recognition of blood group trisaccharides by norovirus. J. Virol. 2007, 81, 5949–5957.
- Chen, R.; Neill, J.D.; Estes, M.K.; Prasad, B.V. X-ray structure of a native calicivirus: Structural insights into antigenic diversity and host specificity. Proc. Natl. Acad. Sci. USA 2006, 103, 8048–8053.
- Tan, M.; Jiang, X. Norovirus and its histo-blood group antigen receptors: An answer to a historical puzzle. Trends Microbiol. 2005, 13, 285–293.
- Tan, M.; Jiang, X. Norovirus gastroenteritis, carbohydrate receptors, and animal models. PLoS Pathog. 2010, 6, e1000983.
- Tan, M.; Jiang, X. Norovirus-host interaction: Multi-selections by human histo-blood group antigens. Trends Microbiol. 2011, 19, 382–388.
- Kroneman, A.; Vega, E.; Vennema, H.; Vinje, J.; White, P.A.; Hansman, G.; Green, K.; Martella, V.; Katayama, K.; Koopmans, M. Proposal for a unified norovirus nomenclature and genotyping. Arch. Virol. 2013, 158, 2059–2068.
- Vega, E.; Barclay, L.; Gregoricus, N.; Shirley, S.H.; Lee, D.; Vinje, J. Genotypic and epidemiologic trends of norovirus outbreaks in the United States, 2009 to 2013. J. Clin. Microbiol. 2014, 52, 147–155.
- Bull, R.A.; White, P.A. Mechanisms of GII.4 norovirus evolution. Trends Microbiol. 2011, 19, 233–240.
- van Beek, J.; Ambert-Balay, K.; Botteldoorn, N.; Eden, J.S.; Fonager, J.; Hewitt, J.; Iritani, N.; Kroneman, A.; Vennema, H.; Vinje, J.; et al. Indications for worldwide increased norovirus activity associated with emergence of a new variant of genotype II.4, late 2012. Euro Surveill. 2013, 18, 8–9.
- Siebenga, J.J.; Lemey, P.; Kosakovsky Pond, S.L.; Rambaut, A.; Vennema, H.; Koopmans, M. Phylodynamic reconstruction reveals norovirus GII.4 epidemic expansions and their molecular determinants. PLoS Pathog. 2010, 6, e1000884.
- Siebenga, J.J.; Vennema, H.; Renckens, B.; de Bruin, E.; van der Veer, B.; Siezen, R.J.; Koopmans, M. Epochal evolution of GGII.4 norovirus capsid proteins from 1995 to 2006. J. Virol. 2007, 81, 9932–9941.
- Noel, J.S.; Fankhauser, R.L.; Ando, T.; Monroe, S.S.; Glass, R.I. Identification of a distinct common strain of "Norwalk-like viruses" having a global distribution. J. Infect. Dis. 1999, 179, 1334–1344.
- Boon, D.; Mahar, J.E.; Abente, E.J.; Kirkwood, C.D.; Purcell, R.H.; Kapikian, A.Z.; Green, K.Y.; Bok, K. Comparative evolution of GII.3 and GII.4 norovirus over a 31-year period. J. Virol. 2011, 85, 8656–8666.
- Mahar, J.E.; Bok, K.; Green, K.Y.; Kirkwood, C.D. The importance of intergenic recombination in norovirus GII.3 evolution. J. Virol. 2013, 87, 3687–3698.
- Wangchuk, S.; Matsumoto, T.; Iha, H.; Ahmed, K. Surveillance of norovirus among children with diarrhea in four major hospitals in Bhutan: Replacement of GII.21 by GII.3 as a dominant genotype. PLoS ONE 2017, 12, e0184826.
- Boonchan, M.; Guntapong, R.; Sripirom, N.; Ruchusatsawat, K.; Singchai, P.; Rungnobhakhun, P.; Tacharoenmuang, R.; Mizushima, H.; Tatsumi, M.; Takeda, N.; et al. The dynamics of norovirus genotypes and genetic analysis of a novel recombinant GII.P12-GII.3 among infants and children in Bangkok, Thailand between 2014 and 2016. Infect. Genet. Evol. 2018, 60, 133–139.
- Liu, J.; Li, S.; Wang, C.; Zheng, L.; Ma, J.; Li, C.; Huo, Y.; Wang, Y. Genomic characterization of GII.3 noroviruses isolated from children in Zhengzhou city, China, 2015/16. Arch. Virol. 2018, 163, 2737–2742.
- Chan, M.C.; Leung, T.F.; Chung, T.W.; Kwok, A.K.; Nelson, E.A.; Lee, N.; Chan, P.K. Virus Genotype Distribution and Virus Burden in Children and Adults Hospitalized for Norovirus Gastroenteritis, 2012–2014, Hong Kong. Sci. Rep. 2015, 5, 11507.
- Li, H.Y.; Zhang, Y.G.; Lei, X.; Song, J.; Duan, Z.J. Prevalence of noroviruses in children hospitalized for acute gastroenteritis in Hohhot, China, 2012–2017. BMC Infect. Dis. 2019, 19, 595.
- Zhou, H.; Wang, S.; von Seidlein, L.; Wang, X. The epidemiology of norovirus gastroenteritis in China: Disease burden and distribution of genotypes. Front. Med. 2020, 14, 1–7.
- Utsumi, T.; Lusida, M.I.; Dinana, Z.; Wahyuni, R.M.; Soegijanto, S.; Soetjipto; Athiyyah, A.F.; Sudarmo, S.M.; Ranuh, R.G.; Darma, A.; et al. Molecular epidemiology and genetic diversity of norovirus infection in children hospitalized with acute gastroenteritis in East Java, Indonesia in 2015-2019. Infect. Genet. Evol. 2021, 88, 104703.
- Satter, S.M.; Abdullah, Z.; Cardemil, C.V.; Flora, M.S.; Gurley, E.S.; Rahman, M.; Talha, M.; Islam, M.D.; Hossain, M.E.; Balachandran, N.; et al. Hospital-based Surveillance for Pediatric Norovirus Gastroenteritis in Bangladesh, 2012–2016. Pediatr. Infect. Dis. J. 2021, 40, 215–219.
- Parker, S.P.; Cubitt, W.D.; Jiang, X. Enzyme immunoassay using baculovirus-expressed human calicivirus (Mexico) for the measurement of IgG responses and determining its seroprevalence in London, UK. J. Med. Virol. 1995, 46, 194–200.
- Zhirakovskaia, E.V.; Tikunov, A.Y.; Bodnev, S.A.; Klemesheva, V.V.; Netesov, S.V.; Tikunova, N.V. Molecular epidemiology of noroviruses associated with sporadic gastroenteritis in children in Novosibirsk, Russia, 2003–2012. J. Med. Virol. 2015, 87, 740–753.
- Saito, M.; Tsukagoshi, H.; Ishigaki, H.; Aso, J.; Ishii, H.; Okayama, K.; Ryo, A.; Ishioka, T.; Kuroda, M.; Saruki, N.; et al. Molecular evolution of the capsid (VP1) region in human norovirus genogroup II genotype 3. Heliyon 2020, 6, e03835.
- Huo, Y.; Chen, X.; Zheng, L.; Huo, J.; Zhang, S.; Wang, M.; Wang, Y. Characterization of virus-like particles derived from a GII.3 norovirus strain distantly related with current dominating strains. Virus Genes 2016, 52, 613–619.
- Song, C.; Takai-Todaka, R.; Miki, M.; Haga, K.; Fujimoto, A.; Ishiyama, R.; Oikawa, K.; Yokoyama, M.; Miyazaki, N.; Iwasaki, K.; et al. Dynamic rotation of the protruding domain enhances the infectivity of norovirus. PLoS Pathog. 2020, 16, e1008619.
- Yang, Y.; Xia, M.; Wang, L.; Arumugam, S.; Wang, Y.; Ou, X.; Wang, C.; Jiang, X.; Tan, M.; Chen, Y.; et al. Structural basis of host ligand specificity change of GII porcine noroviruses from their closely related GII human noroviruses. Emerg. Microbes Infect. 2019, 8, 1642–1657.
- Huo, Y.; Wan, X.; Ling, T.; Shen, S. Biological and immunological characterization of norovirus major capsid proteins from three different genotypes. Microb. Pathog. 2016, 90, 78–83.
- Rockx, B.H.; Vennema, H.; Hoebe, C.J.; Duizer, E.; Koopmans, M.P. Association of histo-blood group antigens and susceptibility to norovirus infections. J. Infect. Dis. 2005, 191, 749–754.
- Hutson, A.M.; Atmar, R.L.; Graham, D.Y.; Estes, M.K. Norwalk virus infection and disease is associated with ABO histo-blood group type. J. Infect. Dis. 2002, 185, 1335–1337.
- Treanor, J.; Sherwood, J.; Cramer, J.P.; Le Cam Bouveret, N.; Lin, S.; Baehner, F.; Borkowski, A.; NOR-204 Investigators. A phase 2 study of the bivalent VLP norovirus vaccine candidate in older adults; impact of MPL adjuvant or a second dose. Vaccine 2020, 38, 5842–5850.
- Sherwood, J.; Mendelman, P.M.; Lloyd, E.; Liu, M.; Boslego, J.; Borkowski, A.; Jackson, A.; Faix, D.; US Navy Study Team. Efficacy of an intramuscular bivalent norovirus GI.1/GII.4 virus-like particle vaccine candidate in healthy US adults. Vaccine 2020, 38, 6442–6449.
- Zhang, M.; Fu, M.; Hu, Q. Advances in Human Norovirus Vaccine Research. Vaccines 2021, 9, 732.
- Richardson, C.; Bargatze, R.F.; Goodwin, R.; Mendelman, P.M. Norovirus virus-like particle vaccines for the prevention of acute gastroenteritis. Expert Rev. Vaccines 2013, 12, 155–167.
- Lindesmith, L.C.; Beltramello, M.; Donaldson, E.F.; Corti, D.; Swanstrom, J.; Debbink, K.; Lanzavecchia, A.; Baric, R.S. Immunogenetic mechanisms driving norovirus GII.4 antigenic variation. PLoS Pathog. 2012, 8, e1002705.
This entry is offline, you can click here to edit this entry!
