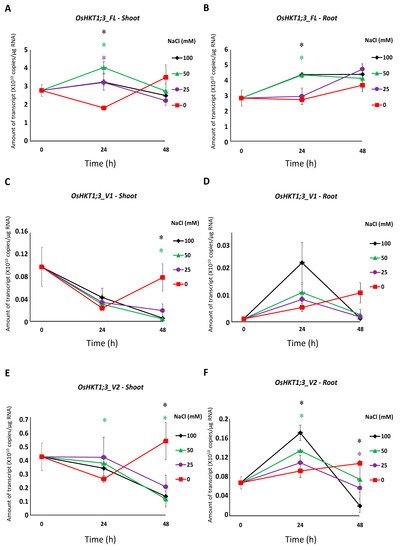
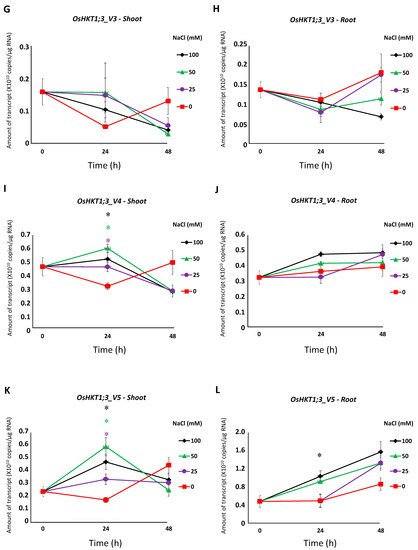
Figure 3. qPCR analyses of
OsHKT1;3 transcripts in Nipponbare seedlings grown in salt-stressed conditions. Expression levels of
OsHKT1;3_FL and each variant of the shoots and roots were investigated by absolute quantification. Fourteen-day-old Nipponbare plants were treated with (control) 0, 25, 50, and 100 mM NaCl solutions for 0, 24, and 48 h prior to total RNA extraction. (
A)
OsHKT1;3_FL-Shoot. (
B)
OsHKT1;3_FL-Root. (
C)
OsHKT1;3_V1-Shoot. (
D)
OsHKT1;3_V1-Root. (
E)
OsHKT1;3_V2-Shoot. (
F)
OsHKT1;3_V2-Root. (
G)
OsHKT1;3_V3-Shoot. (
H)
OsHKT1;3_V3-Root. (
I)
OsHKT1;3_V4-Shoot. (
J)
OsHKT1;3_V4-Root. (
K)
OsHKT1;3_V5-Shoot. (
L)
OsHKT1;3_V5-Root. Absolute amounts of transcripts (copies/μg RNA) are shown. Data are means ± SE,
n = 3. Two independent experiments were performed, and similar results were obtained. An independent
t-test was used to compare the expression. In each variant, data at 24 h from 3 stress conditions (25, 50, and 100 mM NaCl) were subjected to a
t-test vs. control. If a significant difference (
p < 0.05) was detected, such data were marked with an asterisk (*) and colored (purple for 25 mM NaCl, green for 50 mM NaCl, or black for 100 mM NaCl). The same analyses were performed on data at 48 h.
3. Discussion
HKTs play important roles in the salt tolerance, ion homeostasis, and distribution of Na
+ in plant cells and tissues in salt stress conditions, along with other Na
+ transporters [
13,
14,
41].
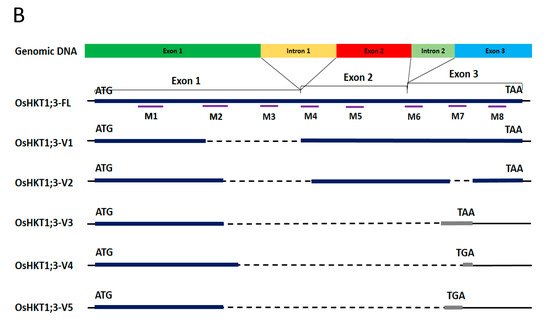
Previously, several splicing variants of
OsHKT1;1 have been reported in the salt-tolerant indica rice, Pokkali [
31]. Similarly to
OsHKT1;1, it has been reported in a Ph.D. thesis from the University of Adelaide that
OsHKT1;3 also produced a spliced variant [
40]. In the present study, several splice variants of
OsHKT1;3 were confirmed in the salt-sensitive japonica rice, Nipponbare (
Figure 1).
Class 1 HKT transporters have been demonstrated to have important roles in Na
+ exclusion and salt tolerance mechanisms in several plant species. In rice, the vital role of
OsHKT1;1 in Na
+ exclusion from the shoots, regulation of Na
+ content in the roots, and the Na
+ recirculation mechanism from the shoots to the roots was demonstrated [
34,
42]. The expression of
OsHKT1;1 increased in the shoots, but not in the roots [
31,
42], in salt stress conditions. OsHKT1;5, a Na
+-selective transporter, has been indicated to protect leaves, including young ones, in rice through Na
+ unloading from the xylem of the roots and sheaths, and the phloem at the basal node, in salt stress conditions [
22,
29,
43].
OsHKT1;4 was demonstrated to be involved in Na
+ exclusion in the stems and leaf sheaths (reducing Na
+ in leaf blades) of a japonica rice cultivar at the reproductive growth stage [
35]. In addition to these
OsHKT1 genes,
OsHKT1;3 was reported to be involved in salt tolerance [
29,
30]. The strong expression of
OsHKT1;3 in bulliform cells, large, highly vacuolated cells of the adaxial epidermis, may indicate the involvement of
OsHKT1;3 in the Na
+ recirculation mechanisms from the shoots to the roots [
29]. However, the detailed physiological functions of
OsHKT1;3 are yet to be elucidated. The present study investigated the function of the
OsHKT1;3 variants in japonica accessions.
OsHKT1;3_FL, identified in the present study, was most-closely similar to the previously registered
OsHKT1;3 (XM_015770707.2) in the National Center for Biotechnology Information (
https://www.ncbi.nlm.nih.gov/nuccore/XM_015770707.2, accessed on 27 July 2021). Oocytes expressing OsHKT1;3_FL in the present study showed a small Na
+ current (
Figure 4A) as reported previously [
30]. However, the transport functions and expressions of
OsHKT1;3 variants have not been investigated so far. As seen in
Figure 4A,B, all oocytes expressing OsHKT1;3 variants showed small currents in the presence of 96 mM Na
+. OsHKT1;3_V3, _V4, and _V5 were short-length variants, and oocytes expressing these variants showed slightly larger bidirectional currents (
Figure 4B), but the biochemical functions and physiological roles of such variants remain to be investigated.
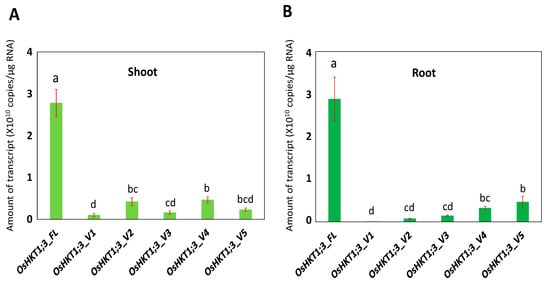
According to Jabnoune et al. [
29], the expression of
OsHKT1;3 showed no significant changes in the roots and leaves in different growth conditions. In addition,
OsHKT1;3 expression levels in the Pokkali variety were lower in the roots than that of the sensitive cultivar, Nipponbare [
37]. Moreover, Farooq et al. [
38] reported recently that
OsHKT1;3 (
OsHKT6) showed high expression in a Cheongcheong rice variety. In the present study, the
OsHKT1;3_FL mRNA was the most abundant in both the roots and shoots among the variants identified (
Figure 2). This was different from the
OsHKT1;1 transcript [
31], in which the transcript of
OsHKT1;1_FL was less abundant, and a variant (
OsHKT1;1_V1) was most abundant.
The expression of OsHKT1;3_FL and some of its variants increased at 24 h of salt stress (Figure 3), and such results may indicate that OsHKT1;3 and its variants were involved in salt tolerance or ion homeostasis at 24 h, at least partially. However, OsHKT1;3 (both FL and all variants) induced only small Na+ currents in the heterologous expression system using X. laevis oocytes (Figure 4), and showed a relatively stable, but not greatly enhanced, expression pattern after NaCl treatment. These results may suggest that OsHKT1;3 mainly played a supplementary role in salt tolerance or a house-keeping role in rice, unlike other OsHKTs that play critical roles in salt tolerance. The present data, together with previous data, have elucidated various characteristics among OsHKT family members, and will provide insights into how they are involved in the mechanisms of ion homeostasis and salt tolerance in rice.