Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
The green peach aphid (Myzus persicae Sulzer), a major and harmful chili aphid usually managed using chemical pesticides, is responsible for massive annual agricultural losses. The efficacy of two protein elicitors, PeaT1 and PeBC1, to stimulate a defensive response against M. persicae in chili was studied in this study.
- PeaT1
- PeBC1
- M. persicae
- induced plant resistance against aphids
1. Introduction
Herbivores and plants have developed a complex interaction over the course of their respective evolutions. Plants are damaged by herbivores and thus modify their physical structures to cope with this damage; as such, plants have developed an economical defense mechanism to protect themselves against herbivores [1]. Herbivores’ development, colonization, feeding, survival, and oviposition are all affected by the structures and substances, which also attract natural adversaries and urge them to develop defence mechanisms [1,2]. Plants have established two primarily constitutive defensive mechanisms to deal successfully with this harm [1]. Plants use physically compromised barriers to resist colonization, such as cuticle trichomes, callose, cell walls, and suberin, but anti-biotic allelochemicals impact or stimulate pest production, fertility, and insect durability [3].
Aphids are phloem-feeding insects that spread plant viruses by syphoning off plant sap, resulting in major agricultural losses [4,5]. Aphid defence responses have been explored in a variety of aphid–plant settings. In green peach aphids M. persicae, Arabidopsis thaliana was shown to be less viable. Infested leaves with Sulzer [6]. Dietary effects were generated in chilli plants, and volatile organic compounds were released, resulting in a repellent effect against infested Bemisia tabaci [7]. Brevicoryne brassicae resistance reduced the survival rate and population growth parameters of immature Plutella xylostella in Brassica napus [8].
Jasmonic acid (JA), salicylic acid (SA), and ethylene stimulate the defensive response in plants (ET) [9]. In plants, salicylic acid (SA) and jasmonic acid (JA) are essential regulators of the induced defensive response [9,10]. The defense against sucking–piercing insects has been associated to SA, while the defense against chewing insects has been linked to JA [11]. ET regulates a variety of defense-related mechanisms in plants [12]. Danaus plexippus promotes JA pathway activation but inhibits SA acquisition in oleander aphids, Aphis nerii; JA has the opposite effect in Asclepias syriaca [12]. Plant responses to herbivory and necrotrophic disease infestations, according to current knowledge, activate the JA and SA defence pathways [10]. Similarly, some elicitors and eliciting components in plants can behave as resistant protein- and nucleotide-binding factors, resulting in aphid resistance [13]. Only a few prior studies have shown that JA and SA have a role in aphid response induction via increased expression of genes including PR-1, PR-2, CHIT1, LOX1, and PAL, all of which have been identified as responses induced by JA–SA after aphid feeding [14,15].
M. persicae Sulzer, a major harmful pest of cucumber, maize, barley, wheat, and beans in China, has a direct impact on crop productivity and quality due to its feeding behaviour. Plant defence responses are triggered by biotic and abiotic elicitors [16]. Elicitors are linked to a variety of diseases, including fungus, bacteria, viruses, and oomycetes. The most prevalent elicitors are proteins, glycoproteins, peptides, lipids, and oligosaccharides [17]. They are divided into two categories: race-specific groups that only elicit a defense response in host plants, and general defence groups that stimulate a defense response in both host and non-host plants [18]. Elicitors are bio factors or chemicals that plants use as signal molecules to promote systemic acquired resistance to diseases or herbivores by activating multiple defensive pathways [19,20]. Microbe-associated molecular patterns (MAMPs) and herbivore-associated molecular patterns (HAMPs) created by herbivorous insect pests are both examples of elicitors. The majority of HAMPs have been identified in pests that are lepidopterous, dipterous, or orthopterous [19]. Volicitin, for example, was discovered in beet armyworms (Spodoptera exigua) as the first herbivore-induced elicitor [21]. Elicitor proteins (MAMPs) from fungal (e.g., Pep-13 and endo-1,4-xylanases from Phytophthora and Trichoderma, respectively) and bacterial pathogens (e.g., flg22 from bacterial flagella) diseases have also been discovered [22,23]. These elicitors play an important role in crop protection because they can induce pest resistance, reduce pest fitness, and limit pest feeding. Elicitors are proteins, glycoproteins, and lipoproteins that activate signalling pathways, the hypersensitive response (HR), and reactive oxygen and nitrogen species (ROS and RNS) responses in plants to generate resistance to diseases and herbivore pests [20,24,25]. Reactive oxygen species (ROS) and nitric oxide (NO), both of which govern metabolic and transcriptional changes, are produced by physiological responses to common processes such as protein phosphorylation or plasma membrane protein activation [20]. Because of the rising demand for food safety, numerous protein elicitors have been investigated as potential pesticide substitutes [26,27,28,29].
PeaT1, broadly specific elicitor examined in Alternaria tenuissima; PeBC1 in Botrytis cinerea and is thought to promote plant resistance via the JA and SA pathways. It activates defense enzymes and strengthens cell walls while also stimulating the production of other defense-related genes [30,31]. Because of their minimal mammalian toxicity and excellent host specificity, entomopathogenic fungi are crucial in the biological control of insect pests [32]. Furthermore, these fungi have the ability to develop as entophytes within various plant parts [32,33]. They also produce systemic resistance in plants against biotic stressors such as phytoparasites, diseases, and nematodes [34], Furthermore, entomopathogenic fungi boost plant development [35], upsurge in the yield [36], and increase the nourishment of plant [37] and the growth of roots [38,39]. Abiotic stresses such as drought [30], iron chlorosis [40], and salinity stress [41] are also mitigated by these fungi. Fungi’s ecological functions have the ability to boost plant health and provide a new perspective on developing novel plant protection techniques [40]. Similarly, certain elicitor proteins from necrotrophic and biotrophic fungal infections have recently been identified, exhibiting induced tolerance to pathogens and herbivores. For example, in A. thaliana, the elicitor PeBC1, which was cloned from the necrotrophic fungus B. cinerea, generated disease resistance [42,43]. PeaT1 (GenBank: EF030819.1) is a type of general elicitor isolated from A. tenuissima. It activates systemic acquired resistance via the SA pathway in plants, resulting in cell wall strengthening and the upregulation of defense-related genes and activation of defense enzymes [44,45]. PeaT1 has been shown to promote growth and strengthen resistance to abiotic stresses in wheat and rice plants [46]. The aim of this study was to look into the activity and molecular mechanism of the B. cinerea-derived elicitor protein PeBC1 and the A. tenuissima-derived elicitor protein PeaT1 in the induction of green peach aphid resistance in chili plants. The impacts of PeaT1 and PeBC1 on M. persicae control, as well as the roles and mechanisms of PeaT1 and PeBC1 on M. persicae control, are investigated in this work to analyze the prospective influence of PeaT1 and PeBC1 on M. persicae. Trichomes were discovered on the leaf’s surface structure, thus prompting researchers to examine the contents of the JA and SA gene expressions from JA and SA. This research also includes information on PeaT1 and PeBC1 function, mechanism, and effects in the integrated management of the green peach aphid (M. persicae).
2. M. persicae Activity
PeaT1 and PeBC1 used two separate strategies to generate resistance to M. persicae, the green peach aphid. First, aphid population fall was stable in PeaT1- and PeBC1-treated chili seedlings (Table 1 and Table 2), with percentage declines in population count in PeaT1 and PeBC1 treatments compared to the buffer and control treatments. M. persicae preferred to feed on the control chili seedlings in the host selection experiment. The frequency of M. persicae colonizing PeaT1- and PeBC1-treated plants was much lower a day after the aphid was inoculated and two days after spraying the seedlings than the control, which revealed aphid colonization in regions other than buffer and PeaT1- and PeBC1-treated areas. Some aphids chose to colonize control regions over those treated with PeaT1 and PeBC1 based on their feeding habits (Figure 1A,B). Second, aphids that were fed on seedlings treated with PeaT1 and PeBC1 had a longer developing period than those that were not, whereas M. persicae that were fed on seedlings treated with PeaT1 and PeBC1 had a lower everyday reproductive ability (second and third nymphal instars). The second and third generations grew at a slower pace (Figure 2A,B).
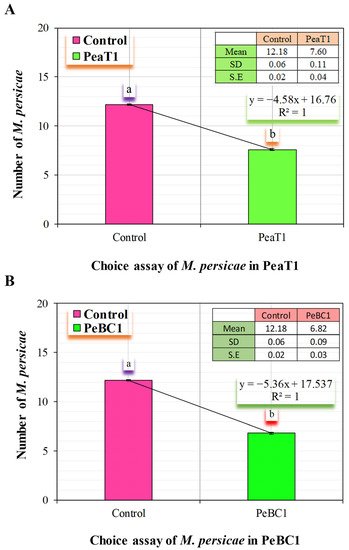
Figure 1. M. persicae feeding preferences (A) M. persicae preferred feeding on control plants in PeaT1; (B) M. persicae feeding on PeBC1-treated and control-treated chilli seedlings 24 h after infestation colonization (mean ± SD). SPSS 18.0 was used to compare data using one-way analysis of variance (ANOVA) and the least significant difference (LSD). Lower style alphabet letters show significant variations across all treatments. (p = 0.05).
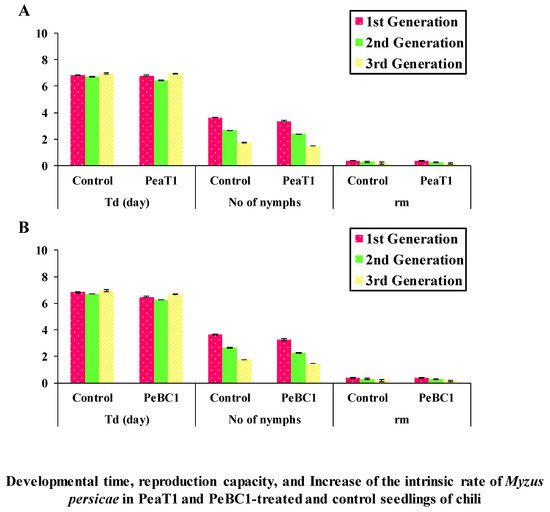
Figure 2. In PeaT1- and PeBC1-treated (A,B) and control seedlings of chili, developmental time, reproductive capacity, and growth of the intrinsic rate of M. persicae are reported as mean ± SD. Td stands for development period, nymphs per day stands for average reproduction ability, and rm stands for intrinsic rate rise. To compare data, SPSS 18.0 was utilized with one-way analysis of variance (ANOVA) and the least significant difference (LSD) (p = 0.05).
Table 1. M. persicae population variations were seen in PeaT1-, control-, and buffer-treated chili seedlings. To compare data, one-way analysis of variance (ANOVA), Levene’s test with SPSS 18.0, and the least significant difference (LSD) were employed. After aphid inoculation on the same day, significant variations in the letters in the rows can be noticed in all treated samples (p = 0.05).
| Days after Aphid Inoculation |
Control | Buffer | PeaT1 |
|---|---|---|---|
| 5 | 53.91 ± 0.05 b | 58.24 ± 0.01 a | 45.23 ± 0.02 c |
| 10 | 108.21 ± 0.04 b | 124.26 ± 0.04 a | 86.24 ± 0.05 c |
| 15 | 214.31 ± 0.06 b | 249.17 ± 0.05 a | 178.26 ± 0.02 c |
Table 2. M. persicae population variations were seen in PeBC1-, control-, and buffer-treated chili seedlings. To compare data, one-way analysis of variance (ANOVA), Levene’s test with SPSS 18.0, and the least significant difference (LSD) were employed. After inoculation of aphid on the same day, significant changes in letters in rows can be noticed in all treated samples (p = 0.05).
| Days after Aphid Inoculation |
Control | Buffer | PeBC1 |
|---|---|---|---|
| 5 | 53.91 ± 0.05 b | 57.40 ± 0.23 a | 43.12 ± 0.03 c |
| 10 | 108.21 ± 0.04 b | 123.35 ± 0.03 a | 84.57 ± 0.04 c |
| 15 | 214.31 ± 0.06 b | 248.15 ± 0.03 a | 175.34 ± 0.06 c |
3. Feeding Activity of M. persicae by EPG
The overall illustration of chili resistance variables was provided by an EPG. M. persicae feeding activity was considerably affected in seedlings treated with PeaT1 and PeBC1 (Table 3 and Table 4). The probing period, the length of C (pathway operation in all tissues), and the sum of M. persicae Pd (potential decrease in cell punctures) in the PeaT1- and PeBC1-treated chilli seedlings were significantly reduced, whereas the period of non-probe time before the first E (phloem-feeding activity) and the total duration of F (penetration problems) increased significantly. During the non-probing period, there was no electrical contact between the aphid stylet and the plant. The non-probing period before the first E was noticeably improved in the PeaT1 and PeBC1 treatments, implying a repellent or deterrent surface feature in the PeaT1 and PeBC1-treated chilli seedlings. C waves show intercellular type motion and may act as a mechanical plant barrier. The shorter the C waves (<3 min) detected, the greater the mechanical difficulty in seedlings treated with PeaT1 and PeBC1. Additionally, a decreased Pd number (cell puncture) was linked to aphid resistance in plants, which could be attributed to mechanical difficulties (the PeaT1- and PeBC1-treated chili seedlings in present study). Wave E1 indicated aphid saliva injection during phloem-feeding activities into sieve elements. In contrast, the E2 wave (sap sucking during phloem-feeding activities) showed phloem sap injection with concurrent salivation, which could have reflected a mesophyll or vascular resistance factor. In the sieve element, an extended E1 indicated more plugging or defense compounds. There was, however, no substantial difference between the control and PeaT1 and PeBC1 treatments in the E2 period, indicating no or low variability in phloem compounds to confer resistance to M. persicae. However, the period of the F wave in the PeaT1- and PeBC1-treated chili seedlings was higher, indicating that PeaT1 and PeBC1 induced an enhanced mechanical defense. The EPG results suggested that the resistance induced by PeaT1 and PeBC1 was mainly due to the modification of physical defenses.
Table 3. M. persicae electrical penetration graph (EPG) data on PeaT1-treated and untreated chilli plants. Mean ± SD. Pathway activities are represented by C, potential drop is represented by Pd, phloem-feeding E represents activities, F represents penetration difficulty, G represents xylem-feeding activities, saliva injection is represented by E1, and sap sucking is represented by E2. Data were compared statistically using an independent t-test with two tails in SPSS 18.0. The difference between PeaT1 and control treatment with the same parameters of * (p = 0.05) is shown by asterisks.
| EPG Parameters | Control (n = 20) | PeaT1 (n = 20) |
|---|---|---|
| Total probing time (h) | 3.78 ± 0.05 | 2.96 ± 0.01 |
| Number of C | 16.45 ± 0.04 | 26.73 ± 0.04 * |
| Number of short probes (C < 3 min) | 9.12 ± 0.07 | 24.12 ± 0.16 |
| Duration of non-probe period before the 1st E (h) | 3.92 ± 0.06 | 3.89 ± 0.06 * |
| Number of pd | 72.87 ± 0.05 | 36.42 ± 0.07 |
| Mean duration of Pd(s) | 8.14 ± 0.04 | 7.69 ± 0.09 |
| Number of E1 | 5.23 ± 0.05 | 4.42 ± 0.07 |
| Mean duration of E1(min) | 8.91 ± 0.04 | 10.12 ± 0.07 |
| Number of E2 | 0.88 ± 0.07 | 0.73 ± 0.07 * |
| Mean duration of E2 (h) | 29.96 ± 0.10 | 43.78 ± 0.05 |
| Number of G | 0.86 ± 0.06 | 0.79 ± 0.08 |
| Mean Duration of G (min) | 19.14 ± 0.04 | 14.67 ± 0.05 |
| Number of F | 5.24 ± 0.05 | 3.13 ± 0.06 |
| mean duration of F (min) | 22.24 ± 0.03 | 52.97 ± 0.04 |
Table 4. M. persicae electrical penetration graph (EPG) data on PeBC1-treated and untreated chilli plants. Mean ± SD. Pathway activities are represented by C, potential drop is represented by Pd, phloem-feeding E represents activities, F represents penetration difficulty, G represents xylem-feeding activities, saliva injection is represented by E1, and sap sucking is represented by E2. Data were compared statistically using an independent t-test with two tails in SPSS 18.0. The difference between PeBC1 and control treatment with the same parameters of * (p = 0.05) is shown by asterisks.
| EPG Parameters | Control (n = 20) | PeBC1 (n = 20) |
|---|---|---|
| Total probing time (h) | 3.14 ± 0.02 | 2.12 ± 0.01 * |
| Number of C | 15.78 ± 0.77 | 25.66 ± 1.61 * |
| Number of short probes (C < 3 min) | 8.67 ± 0.81 | 23.43 ± 1.21 * |
| Duration of non-probe period before the 1st E (h) | 3.76 ± 0.13 | 3.87 ± 0.03 |
| Number of pd | 72.18 ± 0.05 | 35.15 ± 0.03 |
| Mean duration of Pd(s) | 7.71 ± 0.06 | 7.23 ± 0.15 |
| Number of E1 | 4.14 ± 0.02 | 3.45 ± 0.08 |
| Mean duration of E1(min) | 8.79 ± 0.07 | 9.67 ± 0.08 * |
| Number of E2 | 0.64 ± 0.05 | 0.54 ± 0.02 |
| Mean duration of E2 (h) | 29.74 ± 0.02 | 43.27 ± 0.04 |
| Number of G | 0.74 ± 0.06 | 0.68 ± 0.13 |
| Mean Duration of G (min) | 18.23 ± 0.05 | 13.57 ± 0.01 |
| Number of F | 4.67 ± 0.06 | 2.43 ± 0.03 |
| mean duration of F (min) | 21.46 ± 0.02 | ± 0.03 |
This entry is adapted from the peer-reviewed paper 10.3390/microorganisms9112197
This entry is offline, you can click here to edit this entry!
