Towards a comprehensive identification and functional characterization of nutrient-responsive ncRNAs and their downstream molecules, high-throughput sequencing has produced massive omics data for comparative expression profiling as a first step. In this review, we highlight the recent findings of ncRNA-mediated regulation in response to macronutrient stress, with special emphasis on the large-scale sequencing efforts for screening out candidate nutrient-responsive ncRNAs in plants, and discuss potential improvements in theoretical study to provide better guidance for crop breeding practices.
- microRNA
- long noncoding RNA
- circular RNA
- macronutrient
1. Introduction
Nutrient stress is one of the environmental adversities commonly encountered by plants. A thorough understanding of the adaptive strategy to various nutrient stresses will substantially strengthen the theoretical basis for plant breeding practices. Among 14 essential mineral nutrient elements for plant growth and development, six macronutrients, including nitrogen (N), phosphorus (P), potassium (K), calcium (Ca), magnesium (Mg), and sulfur (S), are required in relatively large amounts [1]. Although all these macronutrient elements are mainly absorbed from soil, they differ in sensing and transport pathways, as well as structural and metabolic functions, and may therefore induce different plant responses upon excessive or deficient nutrient supply.
N is the major component of amino acids, nucleotides, chlorophyll, vitamins, and alkaloids, with inorganic ammonium and nitrate being its main form for plant uptake [2]. Both insufficient and excessive levels of N can affect diverse aspects of the plant life cycle. For example, N deficiency could disturb the synthesis of chlorophyll and then reduce the level of photosynthesis, leading to decreased crop yield and quality [3], while an excessive application of N fertilizer might result in a decrease of sugar content [4]. Moderate N deficiency stimulates lateral root growth [5], and severe N deficiency inhibits lateral root growth [6]. P is another essential element involved in vital processes such as photosynthesis, respiration, signal transduction, and nucleic acid synthesis, and it can only be obtained from soil in the form of inorganic phosphorus (Pi) [7,8]. Pi deficiency could not only induce phenotypic changes including dark purple leaf and stem, reduced shoot, and more complex root growth [9], but also cause metabolite alterations such as reduced content of soluble sugar and increased content of organic acids and pigments [10]. K is the third most important macronutrient, and is primarily absorbed by plants in the form of K + . It mainly acts on plants via maintenance of cellular osmotic pressure, adjustment of enzyme activity, optimization of photosynthesis performance, and promotion of assimilation product transport [11]. The impacts of K deficiency on plant yield and quality have also been well documented in a variety of species [12,13,14,15,16,17,18]. Ca is a constituent of cell walls, and mainly participates in maintaining the cell physiological state in plants [19]. It is absorbed by plants in the form of Ca 2+ , a well-known second messenger in cellular signal transduction. A lack of Ca during the fruit ripening process might lead to leakage of cell membranes, irregular softening of cell walls, and abnormal fruit development [20]. Mg is involved in enzyme activation, cell homeostasis, membrane structure stability, active oxygen metabolism, nucleic acid metabolism, and signal transduction [21,22,23]. When Mg deficiency occurs in soil, plant growth is restricted, the leaf becomes yellow, and the biomass allocation between organs changes [24,25,26]. On the other hand, an excess of Mg could result in enhanced and weakened carbon metabolism in roots and leaves, respectively [27]. S is a component of amino acids, sulfated polysaccharides, sulfolipids, and vitamins [28], and plays a decisive role in the structure and biological activity of coenzymes and secondary metabolic products [29]. Plants mainly absorb S from soil in the form of inorganic sulfate through sulfate transporters. S deficiency can also lead to metabolite changes such as increased content of total phenol and reduced content of carotenoid in fruits [30,31].
Among the complex regulatory network underlying plant response to environmental stimuli, noncoding RNAs (ncRNAs) are a class of molecules that play critical roles in coordinating nutrient supply and plant demand. Currently, the most widely studied ncRNAs are microRNAs (miRNAs), which mainly act as negative regulators of their target genes through sequence-specific mRNA cleavage or translational repression [32,33]. In plants, miRNAs have long been known to participate in a wide range of biological processes indispensable for plant growth and stress responses [34,35]. Another subclass of ncRNAs that have been recently recognized as regulators for plant responses to biotic and abiotic stresses are long ncRNAs (lncRNAs) [36], which can either be processed into miRNAs to further modulate the expression of downstream genes, or function as molecular decoys to sequester small RNAs from their target RNAs [37]. In addition, there have also been sporadic reports on the potential involvements of other types of ncRNAs such as circular RNA (circRNA) and cis-natural antisense transcripts (cis-NATs) in nutrient stress response [38,39].
2. miRNA-Mediated Regulation in Response to Macronutrient Stress
Under nutrient stress, miRNA may be either upregulated or downregulated, and hence strengthen or relax its inhibition of target gene expression to adapt to variations in environmental nutrient concentrations. In most cases, the target genes of plant nutrient-responsive miRNAs may encode the sensor or transporter of a certain nutrient element, or the transcription factor in regulation of nutrient homeostasis. In this section, we will summarize major responsive proteins and genes for each macronutrient, with special interests in those targeted by nutrient-responsive miRNAs.
The phosphate starvation response (PHR) is a type of MYB transcription factor that regulates the expression of phosphate-starvation-induced (PSI) genes by binding to the P1BS motif (GNATATNC) in the promoter region. The expression of PHR1 is not sensitive to P starvation while regulated by the SPX-domain protein [60]. In a high P environment, AtSPX1 showed a high binding affinity to AtPHR1, which inhibited the binding of AtPHR1 to the P1BS motif of PSI genes; whereas under low P stress, the affinity between AtSPX1 and AtPHR1 was weakened, and AtPHR1 could activate the expression of PSI genes through binding to their P1BS motifs [61]. Among those PSI genes, the members of SPX-MFS subfamily, including phosphate transporter (PHT) proteins, are involved in intracellular P transport process. In case of P deficiency, the expression of PHT1 was directly induced by PHR1 to promote Pi uptake [62]. Nitrogen limitation adaptation (NLA), another SPX domain protein with E3 ubiquitin ligase activity, can coordinate with the E2 ligase PHO2 to modulate PHT1 degradation, while PHO2 also triggers degradation of PHO1 independent of NLA [63]. PHO1 is an SPX domain protein involved in xylem loading of Pi [64].
The main miRNAs involved in response to P stress are miR399 and miR827 (Figure 1). In Arabidopsis, Ath-miR399 was specifically induced by low P, and it could recognize the AtPHO2 gene and thereby regulate P homeostasis and signaling pathways in plants [65,66,67]. Currently, miR399 has been found in rice, tomato, alfalfa, kidney bean, and strawberry, showing induced expression by P deficiency stress [66,67,68,69]. At the same time, inhibited expression of PHO2 homologous gene by miR399 was also found in rice and kidney bean under low P conditions [70,71]. Interestingly, long-distance movement of miR399s from shoots to roots was discovered in Arabidopsis, and was suggested to be crucial for enhancing Pi uptake and translocation during the onset of Pi deficiency [72]. Later, miR399 and miR395 were also observed to be phloem-mobile in Brassica under nutrient starvation [73]. The function of miR827 was not conserved among different species [74]. Although miR827 was strongly induced under P stress in Arabidopsis and rice, the target gene of Ath-miR827 was AtNLA , while the target of Osa-miR827 was OsPHT5 [75,76]. Similar to miR399, translocation of miR827 and miR2111a between shoots and roots during Pi starvation was also evident in Arabidopsis [77]. In addition, Ath-miR156 and its target, the AtSPL ( squamosa promoter binding protein-like ) gene, were also induced by P stress and participated in stress response by regulating the accumulation of organic acids and anthocyanins in the rhizosphere [78].
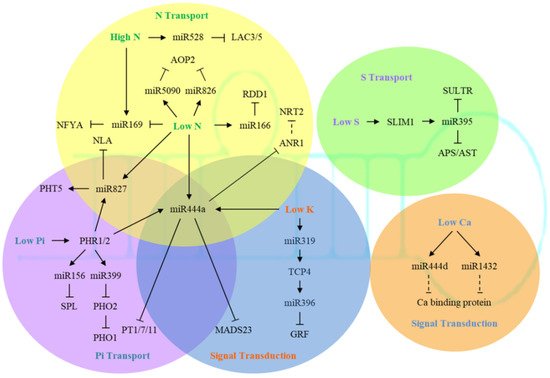
Several studies have identified a large number of K + -transporting proteins in multiple gene families [79]. Among them, AtHAK5 from the KT/KUP/HAK family may function in both low-affinity and high-affinity transport, and is strongly induced by K deficiency [80,81,82,83]. The expression of AtHAK5 was regulated by AP2/ERF transcription factor AtRAP2.11, and could also modulate plant response to low K conditions [84]. So far, there have been 71 K + transporters and channel proteins identified in Arabidopsis. These proteins are not only involved in K uptake and transport in tissues and organs, but also are associated with K storage in vacuoles [85,86,87,88,89].
3. Large-Scale Identification of miRNAs Responsive to Differential Nutrient Availability
Systematic identification of candidate miRNAs involved in plant response to macronutrient stress has been performed in increasing number of species through comparative expression profiling of miRNAs among differentially adapted genotypes or the same genotype under differential nutrient availability, producing large amounts of small RNA sequencing (sRNA-Seq) data (Table 1). Global screening of miRNAs responsive to N starvation was first reported in Arabidopsis, in which miR160, miR780, miR826, miR842, and miR846 exhibited increased expression, while miR169, miR171, miR395, miR397, miR398, miR399, miR408, miR827, and miR857 showed decreased expression upon N stress [57]. Likewise, comparative expression profiling by deep sequencing helped to identified N-responsive miRNAs from shoots and roots of 7-day N-starved rice [50], from leaves and roots of two wheat cultivars subjected to chronic or short-term N stress [109,110], and from shoots and roots of rapeseeds with 0 or 72 h of N-limitation treatment [111].
Table 1. Summary for large-scale identification of macronutrient-stress-responsive miRNAs via high-throughput sequencing.
| Macronutrient Status | Data Quantity | Differentially Expressed miRNAs * | Species/Genotype ** | Tissue | Reference |
|---|---|---|---|---|---|
| N deficiency | 368.1 Mb | 9 ↓, 5 ↑ | Arabidopsis thaliana/Columbia | Seedling | [57] |
| 93.8 Mb | 9 ↓, 13 ↑ | Arabidopsis thaliana/Columbia | Seedling | [112] | |
| 26.6 Gb | 5 ↓, 30 ↑ | Oryza sativa/Nipponbare | Shoot | [50] | |
| 57 ↓, 15 ↑ | Root | ||||
| 1.5 Gb | 2 ↓, 1 ↑ | Triticum turgidum/Svevo | Flag Leaf & Spike | [109,110] | |
| 4 ↓, 5 ↑ | Leaf & Stem | ||||
| 6 ↓, 5 ↑ | Root | ||||
| 1.5 Gb | 2 ↓, 3 ↑ | Triticum turgidum/Ciccio | Flag Leaf & Spike | ||
| 4 ↓ | Leaf & Stem | ||||
| 3 ↓, 4 ↑ | Root | ||||
| 3.4 Gb | 71 ↓, 52 ↑ | Brassica napus /Zhongshuang11 | Shoot | [111] | |
| 64 ↓, 37 ↑ | Root | ||||
| 3.8 Gb | 28 ↓, 8 ↑ | Sorghum bicolor/BTX623 | Shoot | [113] | |
| 25 ↓, 13 ↑ | Root | ||||
| P deficiency | 132.6 Mb | 21 ↑ | Arabidopsis thaliana/Columbia | Shoot | [53] |
| 657.6 Mb | 22 ↓, 33 ↑ | Arabidopsis thaliana/Columbia | Shoot | [75] | |
| 20 ↓, 25 ↑ | Root | ||||
| 961.9 Mb | 27 ↓, 7 ↑ | Glycine max/BX10 | Root | [114] | |
| 40 ↓, 12 ↑ | Shoot | ||||
| 4.3 Gb | 24 ↓, 22 ↑ | Glycine max/Bogao | Root | [115] | |
| 4.4 Gb | 49 ↓, 34 ↑ | Glycine max/Nannong94-156 | Root | ||
| 27.6 Mb | 3 ↓, 2 ↑ | Zea mays/Inbred line 178 | Root | [116] | |
| 14.2 Gb | 174 | Zea mays/Inbred line Q319 | Leaf & Root | [117] | |
| 4.2 Gb | 16 ↓, 33↑ | Sorghum bicolor/BTX623 | Shoot | [113] | |
| 58 ↓, 18↑ | Root | ||||
| K deficiency | 4.4 Gb | 22 ↓, 25 ↑ | Hordeum vulgare/XZ149 | Seedling | [91] |
| 4.5 Gb | 21 ↓, 17 ↑ | Hordeum vulgare/ZD9 | Seedling | ||
| 4.2 Gb | 7 ↓, 5 ↑ | Triticum aestivum/Kenong9204 | Root | [118] | |
| 3.2 Gb | 110 ↓, 122 ↑ | Solanum lycopersicum/JZ18 vs. 35S:SlmiR168a | Leaflet | [119] | |
| 3.8 Gb | 58 ↓, 102 ↑ | Solanum lycopersicum/JZ18 vs. 35S:rSlAGO1 | Leaflet | ||
| 4.0 Gb | 12 ↓, 20 ↑ | Sorghum bicolor/BTX623 | Shoot | [113] | |
| 16 ↓, 6 ↑ | Root | ||||
| Mg deficiency | 2.0 Gb | 71 ↓, 75 ↑ | Citrus sinensis/Xuegan | Leaf | [107] |
| 1.0 Gb | 69 ↓, 101 ↑ | Citrus sinensis/Xuegan | Root | [108] | |
| Ca deficiency | 7.0 Gb | 87 | Arachis hypogea/Baisha1016 | Embryo | [120] |
| S deficiency | 101.9 Mb | 2 ↓, 2 ↑ | Arabidopsis thaliana/Columbia | Seedling | [112] |
The early attempts at large-scale identification of potential P-responsive miRNAs were also reported in Arabidopsis (Table 1), in which the expression of miR156, miR399, miR778, miR827, and miR2111 was induced, but the expression of miR169, miR395, and miR398 was repressed upon P deprivation [53,75]. Since then, an increasing number of candidate P-responsive miRNAs have been obtained in other plant species using different strategies. A microarray-based approach successfully uncovered a subset of 57 known plant miRNAs with differential expression in leaves or roots of soybeans grown under P-deficient and P-sufficient conditions [121], while a genomewide mining dependent on sRNA-Seq identified not only conserved, but also novel miRNAs with significantly altered expression in roots or shoots of a P-efficient genotype soybean treated with low P and high P [114]. Recently, 777 differentially expressed miRNAs across different P treatments and soybean genotypes were also screened out by deep sequencing [115]. Not surprisingly, the sequencing-based expression profiling resulted in substantially larger number of candidate miRNAs than the array-based method did for the same species. In addition, systematic screening of P-responsive miRNAs was also achieved in major crops including rice [122], maize [116,117], and wheat [123].
Global identification of K-deficiency-responsive miRNAs was conducted in roots of two barley genotypes differing in low K tolerance, as well as in wheat roots under five periods of low K treatments, generating approximately 9 Gb and 4 Gb of sRNA-Seq data, respectively ( Table 1 ). The former detected 28 miRNAs differentially expressed at both 2 days and 10 days after low K stress for two barley genotypes [91], while the latter found miR9772, miR1120b-3p, miR531, and miR319 displaying differential expression at all time points during the low K treatments, and suggested that these miRNAs were most possibly involved in mediating plant adaptation to K deficiency [118]. Interestingly, high-throughput sequencing was also employed for identifying differentially expressed miRNAs between two transgenic tomato plants, separately overexpressing SlmiR168a and SlAGO1 , to explore downstream miRNAs (miR171, miR384, miR530, miR858, and miR8007) involved in the SlmiR168 -mediated SlAGO1A regulation upon K stress [119].
Although the basic principle for identifying nutrient-responsive miRNA was to single out candidates with significantly differential expression between or among samples, the calculation method for expression level and the criteria for statistical significance varied among studies. Furthermore, the tissues for sampling also differed among studies, with roots being most frequently used, owing to their high susceptibility to variations in environmental nutrient levels ( Table 1 ). In contrast, there was only one work focusing on the influence of nutrient stress on miRNA abundance in the reproductive tissue of peanut [120]. The lack of progress in achieving nutrient-responsive miRNAs in reproductive tissues might also be attributed to the fact that most of the nutrient-stressed conditions applied in these studies would cause severe symptoms in vegetative tissues and result in failure of flowering or fruit setting. Nevertheless, the results from vegetative tissues still suggested tissue-specific miRNA regulation upon the same type of macronutrient stress. For instance, 13 miRNAs showed similar expression changes in roots and shoots of soybeans under P deficiency, while 6 miRNAs had opposite expression changes in these two tissues [114]. In rapeseed, 11 upregulated and 15 downregulated miRNAs were specifically identified in roots under N starvation, whereas 25 upregulated and 23 downregulated miRNAs were specifically identified in shoots [111].
Another feature of these sequencing efforts for nutrient-responsive miRNA identification
was the integrated analyses of multiomics data. For instance, the downregulation
of miR169 family members, which were identified as N-starvation-responsive miRNAs
in rice based on sRNA-Seq data, could cause the de-repression of NFYA, as validated by
the strand-specific RNA-Seq data [50]. In the same study, the confirmation of MADS25
as a novel target gene of osa-miR444a.4-3p, which was also identified as a N-starvation-responsive miRNA by sRNA-Seq, was aided by analyzing the degradome sequencing data of the N-starved rice [50]. Similarly, combined miRNAome and degradome analysis also helped to reveal the involvement of the miR827-NLA pathway in limited N-induced leaf senescence, as well as the involvement of the miR171-SCL6 and miR160-ARF17 pathways in roots grown under N deprivation [111].
4. Other Types of ncRNAs Involved in Nutrient Stress Response
In addition to stacks of reports on miRNA-mediated regulation during the processes of
plants responding to macronutrient stresses, there are also emerging studies unveiling the
regulatory roles of other types of ncRNAs in these processes, either through interaction with
miRNAs or by directly controlling the expression of protein-coding genes. An early study
in Arabidopsis revealed that lncRNA induced by phosphate starvation 1 (IPS1) contained a
motif complementary to the P-starvation-induced Ath-miR399 with a mismatch loop at the
expected cleavage site, and thus acting as a sponge soaking up Ath-miR399 to inhibit its
cleavage of AtPHO2 transcript [125]. Recently, another lncRNA, T5120, was found to be
modulated by both NLP7 and NRT1.1 to regulate N signaling and improve N use efficiency
in Arabidopsis [126]. On the other hand, a genomewide survey of candidate N-responsive
lncRNAs has been performed through deep sequencing in Populus [127], maize [128],
and barley [129], while lncRNAs responsive to multiple nutrient stresses have also been
explored by deep sequencing in Arabidopsis [130]. Such large-scale sequencing efforts not
only demonstrated an interaction network among lncRNAs, miRNAs, and mRNAs, but
also helped to pinpoint key lncRNAs responsible for nutrient stress response. For instance,
an analysis of RNA-Seq data derived from Arabidopsis roots exposed to low levels of
12 different nutrients revealed trans-acting siRNA3 (TAS3) as a N-responsive lincRNA,
which could produce siRNA targeting NRT2 to regulate N transport and root development
under low N conditions [131]. A recent study based on high-throughput sequencing of
Arabidopsis roots under the treatments of high Ca content or/and a nonpathogenic growth promoting rhizobacterium also proposed a lncRNA–miRNA–mRNA regulatory network
underlying the improved resistance of Arabidopsis to high Ca stress [132].
Another type of ncRNA capable of regulating the function of miRNA and participating
in plant response to macronutrient stress is circRNA, which also serves as an efficient
miRNA sponge [133]. High-throughput sequencing of circRNAs has been conducted
using the leaves harvested from Chinese cabbage at 0, 3, and 6 days after Ca-deficient
treatments [38]. Based on the circRNA-Seq data, dozens of circRNAs with significantly
differential expression in different Ca deficiency stages were isolated, among which one
circRNA was predicted to be a putative sponge for Bra-miR5716 [38]. Likewise, deep
sequencing of the roots from two representative soybean genotypes with different P-use
efficiency also identified 120 differentially expressed circRNAs across different P levels and
genotypes, among which 70 with miRNA-binding sites were suggested as putative miRNA
sponges in response to P deficiency [134]. These circRNA-Seq data will greatly contribute
to elucidating the circRNA-miRNA-mRNA network for nutrient stress response.
One more case to be pointed out is the involvement of cis-NATs, a class of long RNAs
either noncoding or encoding proteins, in plant responses to P stress. In rice, cis-NATPHO1;2
was shown to promote OsPHO1;2 translation without changing the sequence, expression
level, or nuclear export of OsPHO1;2 mRNA, and thereby affecting P homeostasis and
plant fitness [39,125]. However, the detailed regulatory mechanism of cis-NATs in response
to macronutrient stress still needs further study.
5. Conclusions and Perspectives
The roles of ncRNAs, especially miRNAs, in regulating plant responses to nutrient
stress have already been studied for all macronutrient elements, but the responsive
miRNA-target module and the regulatory mechanism vary among elements and species.
Some miRNAs might modulate a crosstalk among multiple nutrient stresses by acting on
different targets. Some miRNAs might also interact with other types of ncRNAs such as
lncRNAs or circRNAs to counteract macronutrient stress. The genomewide expression data
of ncRNAs generated by sRNA-Seq, along with other omics data from the transcriptome,
degradome, proteome, and metabolome, have provided comprehensive insights into the
ncRNA-mediated networks in response to single or multiple nutrient stresses for several
plant species. The candidate nutrient-responsive ncRNAs screened out from these omics
data might serve as promising molecules for further characterization of their detailed functions and for future application in crop genetic engineering. For instance, overexpression of miR5090, which was identified as a N-responsive candidate in Arabidopsis by deep sequencing and subsequent comparative analysis, could lead to improved N uptake and enhanced tolerance to N limitation in transgenic plants [52]. Another case in point was
that the transgenic tobacco lines overexpressing Tae-miR408, a K-deficiency-responsive
candidate in wheat, were also identified from sRNA-Seq data, which showed significantly
improved K uptake, biomass, photosynthesis, and reactive oxygen species scavenging
relative to the wild-type plants under K deficiency [118].
Although global expression profiling through deep sequencing has shown some tissue specific patterns for plant nutrient-responsive ncRNAs, most current studies were based on
vegetative tissues. Considering that reproductive tissues such as fruits and seeds are the primary sources for human diet and animal feed, more research efforts are needed to unravel the impacts of ncRNA-mediated regulation on reproductive tissues under nutrient stress. As aforementioned, such efforts may be hindered by the retarded growth or infertility
owing to the experimental treatments with ultralow macronutrient concentrations that are
not comparable to those in field conditions. Meanwhile, seasonal fluctuations of macronutrient contents in soils are also not neglectable [135]. Therefore, special caution is needed when designing nutrient-stressed treatments for studying plant responses in reproductive tissues. In addition, our preliminary work on the effects of P deficiency on tomato fruit quality also revealed significant alteration in miRNA expression during different stages of fruit development. In this sense, sRNA-Seq data from various developmental stages may enlarge the repertoire of nutrient-responsive ncRNAs and present a spatiotemporal, integrated view of the ncRNA-mediated regulatory network for plant responses to nutrient stress. Taken together, a more precise treatment of nutrient deprivation simulating natural environmental dynamics and a more comprehensive sampling strategy taking reproductive tissues and developmental stages into account will be instrumental in bridging the gap between theoretical study and crop breeding practices.
This entry is adapted from the peer-reviewed paper 10.3390/ijms222011205
