Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Biochemistry & Molecular Biology
Oxidation of membrane lipids by reactive oxygen species (ROS) or O2/lipoxygenase leads to the formation of various bioactive compounds collectively called oxylipins. Reactive carbonyl species (RCS) are a group of oxylipins that have the α,β-unsaturated carbonyl structure, including acrolein and 4-hydroxy-(E)-2-nonenal. RCS provides a missing link between ROS stimuli and cellular responses in plants via their electrophilic modification of proteins.
- RCS
- reactive electrophile species (RES)
- Oxidative stress
- Free radicals
1. Introduction
Plant cells are very rich in reducing agents and redox catalysts. In addition, the above-ground parts contain many pigments. Such conditions are favorable for the production of reactive oxygen species (ROS) via the reduction or excitation of O2, and indeed, there are large fluxes of ROS production [1]. If their levels are regulated properly, then ROS are excellent molecules for biological signals; they can turn on biochemical switches temporally in micro-local regions in a cell, and due to their lifetime, the signal is turned off in a short time.
One of the earliest reports of ROS signaling in plants was on the systemic defense response, demonstrating H2O2 as a remote signal to bring the oxidative information emitted from the local photooxidized leaves [2]. ROS act as signals, not only in stress responses, but also in hormonal responses and development [3]. It has been shown that the abscisic acid (ABA)-induced activation of respiratory burst oxidase-homolog NADPH oxidases (RBOH) in guard cells contributes to the stomata closure response through the action of H2O2 [4]. Involvement of ROS has also been suggested in the auxin-induced formation of lateral roots [5] and adventitious roots [6]. ROS are also critical signals for initiating programmed cell death (PCD) in development [7], senescence and pathogen response [8]. Today, there is keen interest in the biochemical processes of ROS signal transduction in plants, but the signaling pathway from ROS to putative sensor/receptor proteins has been poorly elucidated.
2. Response of Plants to Exogenously Added RCS
2.1. Reaction of RCS with Proteins
Carbonyl compounds are electrophiles and can react with nucleophilic groups on biomolecules to make a covalent bond [9]. Typical reactions are Schiff base formation and Michael addition. Schiff base is formed between a carbonyl group and an amino group through dehydration (Figure 3A). This reaction proceeds fast at pH 5, but slower at lower or higher pH conditions. The formed Schiff base can be hydrolyzed under a highly acidic condition. When a thiol group or an amino group reside nearby the Schiff base, the secondary reactions may proceed to form a complex structure [9], which may contribute to the formation of cross-link between proteins. Michael addition is the reaction of the electrophilic β carbon of an RCS molecule with the thiol sulfur, amino nitrogen, or the imidazole τ-nitrogen atom to form a covalent bond (Figure 3B). They also react with the guanine base of nucleic acids, leading to a mutation [10]. The Michael reaction with a thiol is reversible. The equilibrium constant of the reaction between GSH and an RCS differs greatly, depending on the type of RCS molecules [11].
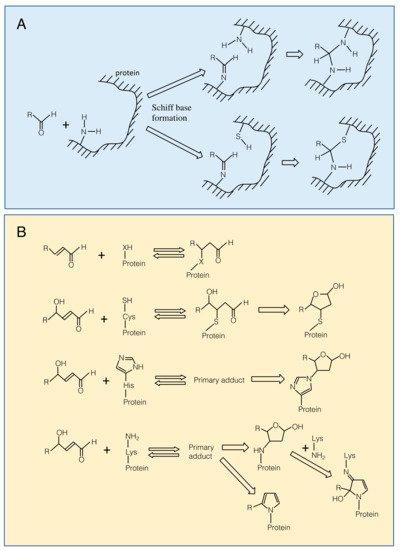
Figure 3. Modification of amino acid residues on a protein with carbonyls. Panel A. Schiff base formation on an amino group with an aldehyde and possible secondary reactions. Panel B. Top, Michael addition of an RCS to a nucleophilic (-XH) group. X is for sulfur in Cys, ε-amino nitrogen in Lys and imidazole τ-nitrogen in His residues. From 2nd to bottom, Michael addition of a 4-hydroxy-(E)-2-alkenal to Cys, His and Lys residues. Primary adducts may undergo cyclization.
Michael addition of an RCS to a protein adds a carbonyl moiety on the protein (protein carbonylation), while Schiff base formation of RCS with a protein does not. Carbonyl moieties on a protein can also be formed by ROS directly at Trp, His, Tyr, Met and Cys residues [12][13]. Protein carbonylation has been conventionally regarded as an indicator of protein oxidation in plants, but it should be noted that a considerable portion of the detected proteins as ‘oxidized proteins’ are modified with RCS [14].
The primary Michael adducts, or protein carbonyls, can proceed to secondary reactions with another nucleophilic group, to produce irreversible structural changes on the protein (Figure 3B). The primary Michael adduct between a nucleophile and 4-hydroxyl-2-alkenals, e.g., HNE and HHE, can undergo cyclization. The cyclic secondary products may further react with another nucleophilic group [10]. These reactions can lead to the formation of cross-links between proteins.
Effects of RCS modification on the protein function differs by the kinds of RCS and proteins. The RCS includes carbonyls with various structure; different carbon chain lengths, number of unsaturated bonds, and the extent of oxygenation [15][16]. Hence, every RCS has different properties such as polarity, hydrophobicity/hydrophilicity, solubility, and volatility [17]. This variety gives distinct RCS molecules different reactivity with proteins, implying that each species can bring distinct information via specific recognition by the targets and scavenging enzymes. For example, HNE and 4-oxo-(E)-2-nonenal (ONE), two C9-RCS differing only by the extent of oxidation at the 4th carbon, show a great difference in the reactivity with GSH; the second-order reaction constant of the former is 100-fold smaller than that of the latter [18]. Interestingly, these two RCS react with the redox sensor protein mitoNEET differently, but in a manner not simply deduced from the reactivity discussed above; ONE is bound to Lys55 specifically, while HNE adds to Lys and His residues broadly on the protein [19]. Several isozymes of glutathione transferase (GST) Tau class recognize acrolein and HNE as substrates, but there are ones that accept acrolein only [20][21]. These examples suggest the difficulty in generalizing the mode of interaction between RCS and their target proteins.
Lists of proteins that are affected by RCS are available for animal proteins [22][23][24], but only a few plant proteins have been investigated, as described below. In general, when an RCS acts as a signal, the (putative) target protein probably will gain the function, so that modification of a small portion of the total population of the target might trigger the next signal. In case of damage, inactivation of an RCS target protein will affect a cellular process immediately if the activity of that protein limits the metabolic or signaling pathway involving it. Extensive modification of a protein will also facilitate cross-linking with other proteins, leading to aggregation unless the modified proteins are proteolytically removed (described below). Under severe and prolonged oxidative stress, larger populations of broader range of proteins will be inactivated, leading to extensive deterioration of cell functions.
Plant proteins that are sensitive to or modified by RCS are listed. Addition of HNE to isolated mitochondria caused sensitive inactivation of lipoate enzymes such as H-subunit (SU) of glycine decarboxylase complex (GDC) [25] and alternative oxidase [26]. Addition of RCS, especially acrolein, to chloroplasts caused a rapid consumption of GSH, followed by the inactivation of phosphoribulokinase (PRK) preferentially, then fructose-1,6-bisphosphatase, glyceraldehyde-3-phosphate dehydrogenase, leading to the loss of the CO2 fixation ability [27]. By a proteomic analysis of A. thaliana cultured cells treated with oxidative agents such as H2O2, menadione and antimycin A, subsets of inner membrane proteins and matrix proteins (totally 31 different proteins) were identified as HNE targets because they were modified by this RCS to greater extents under the stress [28]. In heat-stressed spinach leaves, OEC33 protein in photosystem II (PSII) was modified with MDA and acrolein, while in A. thaliana leaves the antenna LHCII protein was sensitively modified with MDA in heat stress [29]. Mano et al. [14] analyzed the RCS-modification of soluble proteins in leaves from salt stressed A. thaliana plants. As detected with specific antibodies, protein modification with HNE, 4-hydroxy-(E)-2-hexenal, acrolein, crotonaldehyde and malondialdehyde increased in leaves with the progress of the salt-stress treatment. In addition, the acrolein- and crotonaldehyde-modifications were increased significantly even under less severe stress conditions, in which there was no apparent tissue injury or the photoinhibition of PSII. The band pattern of Western blotting suggested these different RCS targeted a common set of proteins. With a quantitative proteomic analysis after immuno-affinity trapping of the HNE-modified proteins, 17 distinct proteins were identified as sensitive targets. Interestingly, these target proteins were distributed to various cellular compartments, i.e., cytoplasm, peroxisome, chloroplast, nucleus and even apoplast. Addition of acrolein to tobacco bright yellow-2 (BY-2) cultured cells or the cell extract activated caspase-1-like protease (C1LP) and caspase-3-like protease (C3LP) activities [30]. Results of these experiments with different samples and treatments commonly suggest that certain subsets of proteins are sensitively modified with RCS under stress conditions. In other words, intracellular RCS do not necessarily react with broad range of proteins.
The fate of RCS-modified proteins has not been significantly investigated, both in animals and plants. In general, ‘damaged’ proteins should be immediately degraded before extensive modification leads to the formation of aggregated proteins, which may escape degradation and deposit in cells to cause detrimental effects. Mildly modified proteins are degraded by proteasomes [31]. In mice, the 20S proteasome has been shown to be responsible for the elimination of most oxidized proteins [32]. In A. thaliana also, the 20S proteasome appears to be responsible for the degradation of oxidized proteins, while the 26S proteasome takes care of misfolded proteins [33]. In mitochondria, ATP-dependent proteases such as Lon protease has been recognized as key proteases for the removal of oxidized proteins in mammals and yeast, but not in plants [34]. These studies have investigated “oxidized” or “carbonylated” proteins collectively and have not distinguished ROS-mediated and RCS-mediated modification on proteins. There is a report that the degradation HNE-modified protein is catalyzed by cathepsin G in rat [35], but corresponding facts for plants are not available. It should be noted that the modification of proteins with certain kinds of RCS such as MDA are reversible [36]. Thus, it is possible that a transient increase in the RCS concentration modifies the target protein(s) and afterwards the RCS is released from the protein, and then scavenged by certain enzymes (described below). In this case the target protein can be recruited to its physiological role although there has been no experimental data available for demonstrating such dynamics.
2.2. Cytotoxicity
RCS added from outside of the cells are toxic to plants when their concentrations are high. Reynolds [37] evaluated the toxicity of various carbonyls for their ability to inhibit germination of lettuce seeds and demonstrated that RCS have relatively higher toxicity than non-RCS carbonyls. Exposure of A. thaliana plants to acrolein or methylvinylketone (MVK) as volatiles (10 μmol/L air) caused a decrease in PSII activity in 15 h [38]. Similarly, (E)-2-hexenal (100 μmol/L air) significantly decreased PSII activity in 5 h [39]. When a 20 μM aqueous solution of HNE was infiltrated into A. thaliana leaves, PSII was inactivated in 15 h [38] and higher concentrations at mM levels caused necrosis [40]. Aqueous 10 μM HNE inhibited root elongation in tobacco plants [41].
At subcellular levels, the addition of HNE to isolated mitochondria inactivated respiratory metabolism in the matrix [42]. In this case, the most sensitive targets were lipoate enzymes as described above [25]. When isolated chloroplasts were treated with acrolein in darkness, their photosynthetic activity was lost, but photosynthetic electron transport chain was insensitive [27]. On the other hand, the addition of acrolein to the Synechocystis cells under light inactivated PSII and caused growth inhibition. The inactivation mechanism is accounted for by the combined effect of acrolein and hydroxyl radical, which was generated under light [43].
2.3. Signaling Effects of Exogenously Added RCS
Exogenous application of RCS at low levels to whole plants can induce arrays of defense genes. In A. thaliana seedlings exposed to (E)-2-hexenal, a C6 volatile RCS that can be produced enzymatically in response to pathogen attack, defense response genes including phenylpropanoid synthesis enzymes were induced [44][45]. Low levels of MDA strongly upregulated many abiotic/environmental stress-related genes such as ROF1 and XERO2 in A. thaliana [36]. When acrolein and MVK were infiltrated to A. thaliana leaves, pathogenesis-related genes such as HEL/PR4 were activated [38]. Fumigation of A. thaliana plants with RCS induced a group of heat shock response genes [46]. The putative RCS receptor involved in this response should have a specificity to certain types of RCS because this signaling effect was observed for RCS of carbon chain length 4–8, but not for the C3 RCS acrolein. Addition of acrolein, HHE and HNE to A. thaliana caused increases in the activity of various ROS scavenging enzymes such as catalase and ascorbate peroxidase [47]. Interestingly, the extreme halophyte Eutrema parvulum, a close relative to A. thaliana, did not respond similarly to these RCS; the enzyme activities were not increased. This suggests that the RCS perception systems or RCS scavenging selectivity and capacity in halophytes are different from those in glycophytes.
The addition of acrolein to C. reinhardtii cells up to 600 ppm increased the GSTS1 content and improved cells’ tolerance to 1O2. On the basis of transcriptomic analysis, it is suggested that acrolein can mediate the gene expression signal triggered by 1O2 [48].
OPDA, a phytohormone and a precursor to jasmonic acid (JA), has a cyclopentenone structure and is an RCS. When applied to A. thaliana plants exogenously, OPDA induces a set of defense genes that is distinct from those induced by JA [49], and represses the expression of those involved in cell cycle regulation and cell growth [50]. This OPDA signal is recognized by the TGA transcription factors [51]. OPDA can be bound also to cyclophilin 20-3 and alter the protein’s ability to trigger the formation of cysteine synthase complex, and thereby affect the redox homeostasis [52].
This entry is adapted from the peer-reviewed paper 10.3390/plants8100391
References
- Foyer, C.H.; Noctor, G. Redox sensing and signalling associated with reactive oxygen in chloroplasts, peroxisomes and mitochondria. Physiol. Plant. 2003, 119, 355–364.
- Karpinski, S.; Reynolds, H.; Karpinska, B.; Wingsle, G.; Creissen, G.; Mullineaux, P. Systemic signaling and acclimation in response to excess excitation energy in Arabidopsis. Science 1999, 284, 654–657.
- Mittler, R. ROS are good. Trends Plant Sci. 2017, 22, 11–19.
- Pei, Z.M.; Murata, Y.; Benning, G.; Thomine, S.; Klüsener, B.; Allen, G.J.; Grill, E.; Schroeder, J.I. Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 2000, 406, 731–734.
- Orman-Ligeza, B.; Parizot, B.; de Rycke, R.; Fernandez, A.; Himschoot, E.; Breusegem, F.V.; Bennett, M.J.; Périlleux, C.; Beeckman, T.; Draye, X. RBOH-mediated ROS production facilitates lateral root emergence in Arabidopsis. Development 2016, 143, 3328–3339.
- Li, S.; Xue, L.; Xu, S.; Feng, H.; An, L. Hydrogen peroxide involvement in formation and development of adventitious roots in cucumber. Plant Growth Regul. 2007, 52, 173–180.
- Bethke, P.C.; Jones, R.L. Cell death of barley aleurone protoplasts is mediated by reactive oxygen species. Plant J. 2001, 25, 19–29.
- Torres, M.A.; Jones, J.D.G.; Dangl, J.L. Pathogen-induced, NADPH oxidase–derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat. Genet. 2005, 37, 1130–1134.
- Schauenstein, E.; Esterbauer, H.; Zollner, H. Aldehyde in Biological Systems: Their Natural Occurrence and Biological Activities; Pion: London, UK, 1977; pp. 589–590.
- Esterbauer, H.; Schauer, R.; Zollner, J.H. Chemistry and biochemistry of 4-hydroxynonenal, malondialdehyde and related aldehydes. Free Rad. Biol. Med. 1991, 11, 81–128.
- Esterbauer, H.; Zollner, H.; Scholtz, N. Reaction of glutathione with conjugated carbonyls. Zeitschrift für Naturforschung C 1975, 30, 466–473.
- Møller, I.M.; Jensen, P.E.; Hansson, A. Oxidative modifications to cellular components in plants. Annu. Rev. Plant Biol. 2007, 58, 459–481.
- Bachi, A.; Dalle-Donne, I.; Scaloni, A. Redox proteomics: Chemical principles, methodological approaches and biological/biomedical promises. Chem. Rev. 2013, 113, 596–698.
- Mano, J.; Nagata, M.; Okamura, S.; Shiraya, T.; Mitsui, T. Identification of oxidative-modified proteins in salt-stressed Arabidopsis: A carbonyl-targeted proteomics approach. Plant Cell Physiol. 2014, 55, 1233–1244.
- Mano, J. Reactive carbonyl species: Their production from lipid peroxides, action in environmental stress, and the detoxification mechanism. Plant Physiol. Biochem. 2012, 59, 90–97.
- Yalcinkaya, T.; Uzilday, B.; Ozgur, R.; Turkan, I.; Mano, J. Lipid peroxidation-derived reactive carbonyl species (RCS): Their interaction with ROS and cellular redox during environmental stresses. Environ. Exp. Bot. 2019, 165, 139–149.
- LoPachin, R.M.; Gavin, T.; Peterson, D.R.; Barber, D.S. Molecular mechanisms of 4-hydroxyn-2-nonenal and acrolein toxidity: Nucleophilic targets and adduct formation. Chem. Research Toxicol. 2009, 22, 1499–1508.
- Rudolph, T.K.; Freeman, B.A. Transduction of redox signaling by electrophile-protein reactions. Sci. Signal. 2009, 2, re7.
- Arnette, D.; Quillin, A.; Gledenhuys, W.J.; Menze, M.A.; Konkle, M. 4-Hydroxynonenal and 4-ocononenal differentially bind to the redox sensor mitoNEET. Chem. Res. Toxicol. 2019, 32, 977–981.
- Mano, J.; Kanameda, S.; Kuramitsu, R.; Matsuura, N.; Yamauchi, Y. Detoxification of reactive carbonyl species by glutathione transferase Tau isozymes. Front. Plant Sci. 2019, 10, 487.
- Mano, J.; Ishibashi, A.; Muneuchi, H.; Morita, C.; Sakai, H.; Biswas, S.; Kitajima, S. Acrolein-detoxifying isozymes of glutathione transferase in plants. Planta 2017, 245, 255–264.
- Wible, R.S.; Sutter, T.R. Soft cysteine signaling network: The functional significance of cysteine in protein function and the soft acids/bases thiol chemistry that facilitates cysteine modification. Chem. Res. Toxicol. 2017, 30, 729–762.
- Schopfer, F.J.; Cipollina, C.; Freeman, B.A. Formation and signaling actions of electrophilic lipids. Chem. Rev. 2011, 111, 5997–6021.
- Pampola, R. Advanced lipoxidation end-products. Chem. Biol. Interact. 2011, 192, 14–20.
- Taylor, N.L.; Day, D.A.; Millar, A.H. Environmental stress causes oidative damage to plant mitochondria leading to inhibition of glycine decarboxylase. J. Biol. Chem. 2002, 277, 42662–42668.
- Winger, A.M.; Millar, A.H.; Day, D.A. Sensitivity of plant mitochondrial terminal oxidases to the lipid peroxidation product 4-hydroxy-2-nonenal (HNE). Biochem. J. 2005, 387, 865–870.
- Mano, J.; Miyatake, F.; Hiraoka, E.; Tamoi, M. Evaluation of the toxicity of stress-related aldehydes to photosynthesis in chloroplasts. Planta 2009, 230, 639–648.
- Winger, A.M.; Taylor, N.L.; Heazlewood, J.L.; Day, D.A.; Miller, A.H. The cytotoxic lipid peroxidation product 4-hydroxy-2-nonenal covalently modifies a selective range of proteins linked to respiratory function in plant mitochondria. J. Biol. Chem. 2007, 282, 37436–37447.
- Yamauchi, Y.; Furutera, A.; Seki, K.Y.; Toyoda, Y.; Tanaka, K.; Sugimoto, Y. Malondialdehyde generated from peroxidized linoleic acid causes protein modification in heat-stressed plants. Plant Physiol. Biochem. 2008, 46, 786–793.
- Biswas, M.S.; Mano, J. Reactive carbonyl species activate caspase-3-like protease to initiate programmed cell death in plants. Plant Cell Physiol. 2016, 57, 1432–1442.
- Viestra, R.D. The ubiquitin/26S proeasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci. 2003, 8, 135–142.
- Zheng, J.; Bizzozero, O. Reduced proteasomal activity contributes to the accumulation of carbonylated proteins in chromic experimental autoimmune encephalomyelitis. J. Neurochem. 2010, 115, 1556–1567.
- Kurepa, J.; Toh-e, A.; Smalle, J.A. 26S proteasome regulatory particle mutants have increased oxidative stress tolerance. Plant J. 2008, 53, 102–114.
- Smakowska, E.; Czarna, M.; Janska, H. Mitochondrial ATP-dependent proteases in protection against accumulation of carbonylated proteins. Mitochondrion 2014, 19B, 245–251.
- Tsuchiya, Y.; Okada, G.; Kobayashi, S.; Chikuma, T.; Hojo, H. 4-Hydroxy-2-nonenal-modified glyceraldehyde-3-phosphate dehydrogenase is degraded by cathepsin G in rat neutrophils. Oxid. Med. Cell. Longev. 2011, 2011, 213686.
- Weber, H.; Chételat, A.; Reymond, P.; Farmer, E.E. Selective and powerful stress gene expression in Arabidopsis in response to malondialdehyde. Plant J. 2004, 37, 877–888.
- Reynolds, T. Comparative effects of aliphatic compounds on inhibition of lettuce fruit germination. Ann. Bot. 1977, 41, 637–648.
- Alméras, E.; Stolz, S.; Vollenweider, S.; Reymond, P.; Mène-Saffrané, L.; Farmer, E.E. Reactive electrophile species activate defense gene expression in Arabidopsis. Plant J. 2003, 34, 205–216.
- Matsui, K.; Sugimoto, K.; Mano, J.; Ozawa, R.; Takabayashi, J. Differential metabolism of green leaf volatiles in injured and intact parts of a wounded leaf meet distinct ecophysiological requirements. PLoS ONE 2012, 7, e36433.
- Mano, J.; Belles-Boix, E.; Babiychuk, E.; Inzé, D.; Torii, Y.; Hiraoka, E.; Takimoto, K.; Slooten, L.; Asada, K.; Kushnir, S. Protection against photooxidative injury of tobacco leaves by 2-alkenal reductase. Detoxication of lipid peroxide-derived reactive carbonyls. Plant Physiol. 2005, 139, 1773–1783.
- Yin, L.; Mano, J.; Wang, S.; Tsuji, W.; Tanaka, K. The involvement of lipid peroxide-derived aldehydes in aluminum toxicity of tobacco roots. Plant Physiol. 2010, 152, 1406–1417.
- Millar, A.H.; Leaver, C.L. The cytotoxic lipid peroxidation product, 4-hydroxy-2-nonenal, specifically inhibits decarboxylating dehydrogenases in the matrix of plant mitochondria. FEBS Lett. 2000, 481, 117–121.
- Shimakawa, G.; Iwamoto, T.; Mabuchi, T.; Saito, R.; Yamamoto, H.; Amako, K.; Sugimoto, T.; Makino, A.; Miyake, C. Acrolein, an α,β-unsaturated carbonyl, inhibits both growth and PSII activity in the cyanobacterium Synechocystis sp. PCC 6803. Biosci. Biotechnol. Biochem. 2013, 77, 1655–1660.
- Kishimoto, K.; Matsui, K.; Ozawa, R.; Takabayashi, J. Volatile C6-aldehydes and allo-ocimene activate defense genes and induce resistance against Botrytis cinerea in Arabidopsis thaliana. Plant Cell Physiol. 2005, 47, 1093–1102.
- Bate, N.J.; Rothstein, S.J. C6-volatiles derived from the lipoxygenase pathway induce a subset of defense-related genes. Plant J. 1998, 16, 561–569.
- Yamauchi, Y.; Kunishima, M.; Mizutani, M.; Sugimoto, Y. Reactive short-chain leaf volatiles act as powerful inducers of abiotic stress-related gene expression. Sci. Rep. 2015, 5, 8030.
- Yalcinkaya, T.; Uzilday, B.; Ozgur, R.; Turkan, I. The roles of reactive carbonyl species in induction of antioxidant defence and ROS signalling in extreme halophytic model Eutrema parvulum and glycophytic model Arabidopsis thaliana. Exp. Environ. Bot. 2019, 160, 81–91.
- Roach, T.; Stöggl, W.; Baur, T.; Kranner, I. Distress and eustress of reactive electrophiles and relevance to light stress acclimation via stimulation of thiol/disulfide-based defences. Free Rad. Biol. Med. 2018, 122, 65–73.
- Taki, N.; Sasaki-Sekimoto, Y.; Obayashi, T.; Kikuta, A.; Kobayashi, K.; Ainai, T.; Yagi, K.; Sakurai, N.; Suzuki, H.; Masuda, T.; et al. 12-Oxo-phytodienoic acid triggers expression of a distinct set of genes and plays a role in wound-induced gene expression in arabidopsis. Plant Physiol. 2005, 139, 1268–1283.
- Eckardt, N.A. Oxylipin signaling in plant stress responses. Plant Cell 2008, 20, 495–497.
- Stotz, H.U.; Mueller, S.; Zoeller, M.; Mueller, M.J.; Berger, S. TGA transcription factors and jasmonate-independent COI1 signalling regulate specific plant responses to reactive oxylipins. J. Exp. Bot. 2013, 64, 963–975.
- Park, S.-W.; Li, W.; Viehhauser, A.; He, B.; Kim, S.; Nilsson, A.K.; Andersson, M.X.; Kittle, J.D.; Ambavaram, M.M.R.; Luan, S.; et al. Cyclophilin 20-3 relays a 12-oxo-phytodienoic acid signal during stress responsive regulation of cellular redox homeostasis. Proc. Natl. Acad. Sci. USA 2013, 110, 9559–9564.
This entry is offline, you can click here to edit this entry!
