CELF (CUGBP Elav-like family) proteins are RBPs (RNA-binding proteins) with pleiotropic capabilities in RNA processing. Their responsibilities extend from alternative splicing and transcript editing in the nucleus to mRNA stability, and translation into the cytoplasm. In this way, CELF family members have been connected to global alterations in cancer proliferation and invasion, leading to their identification as potential tumor suppressors or even oncogenes. Notably, genetic variants, alternative splicing, phosphorylation, acetylation, subcellular distribution, competition with other RBPs, and ultimately lncRNAs, miRNAs, and circRNAs all impact CELF regulation.
- CELF proteins
- RNA-binding protein
- noncoding RNA
- cancer
1. Introduction
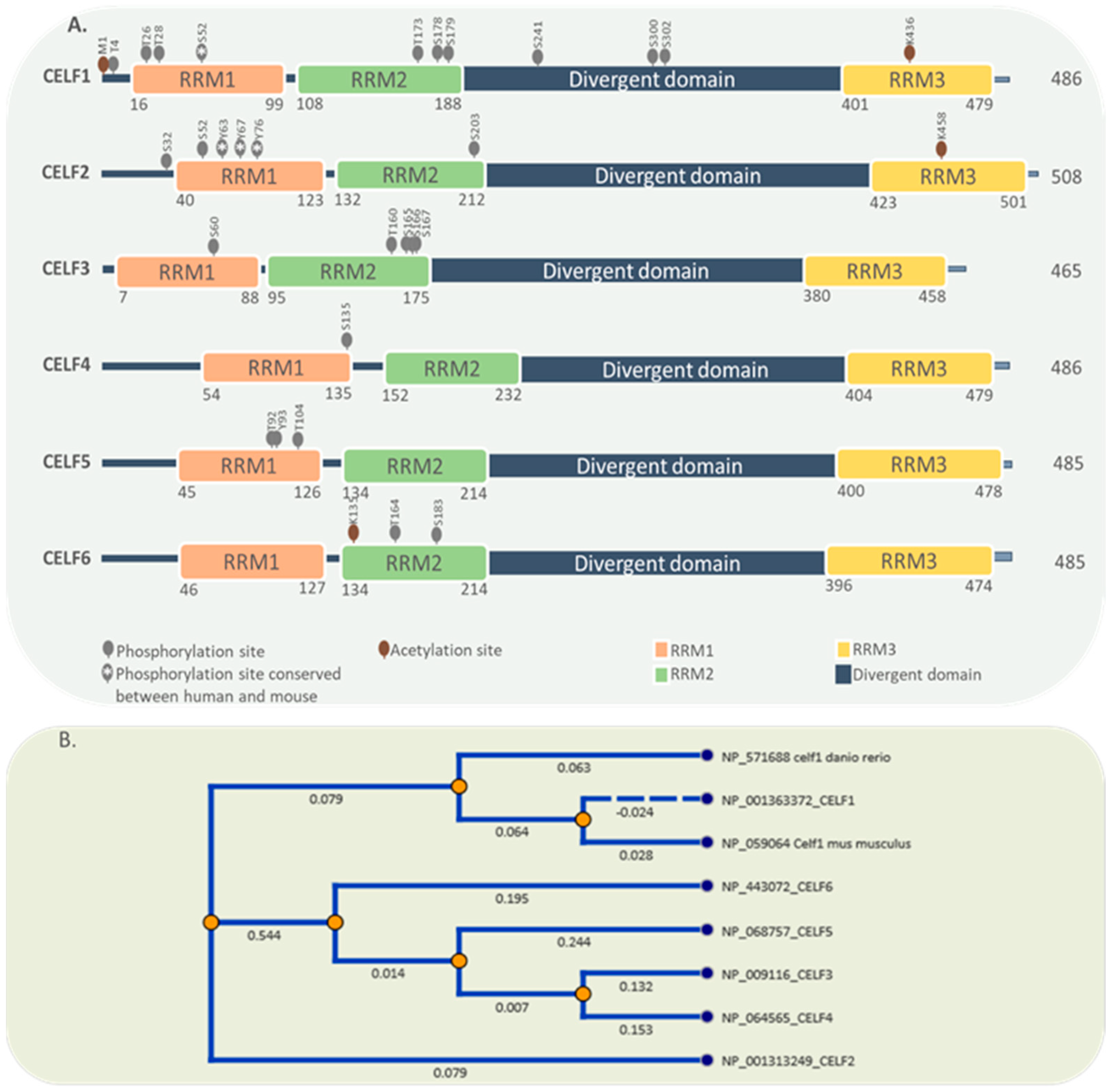
2. General Characteristics of the CELF Protein Family
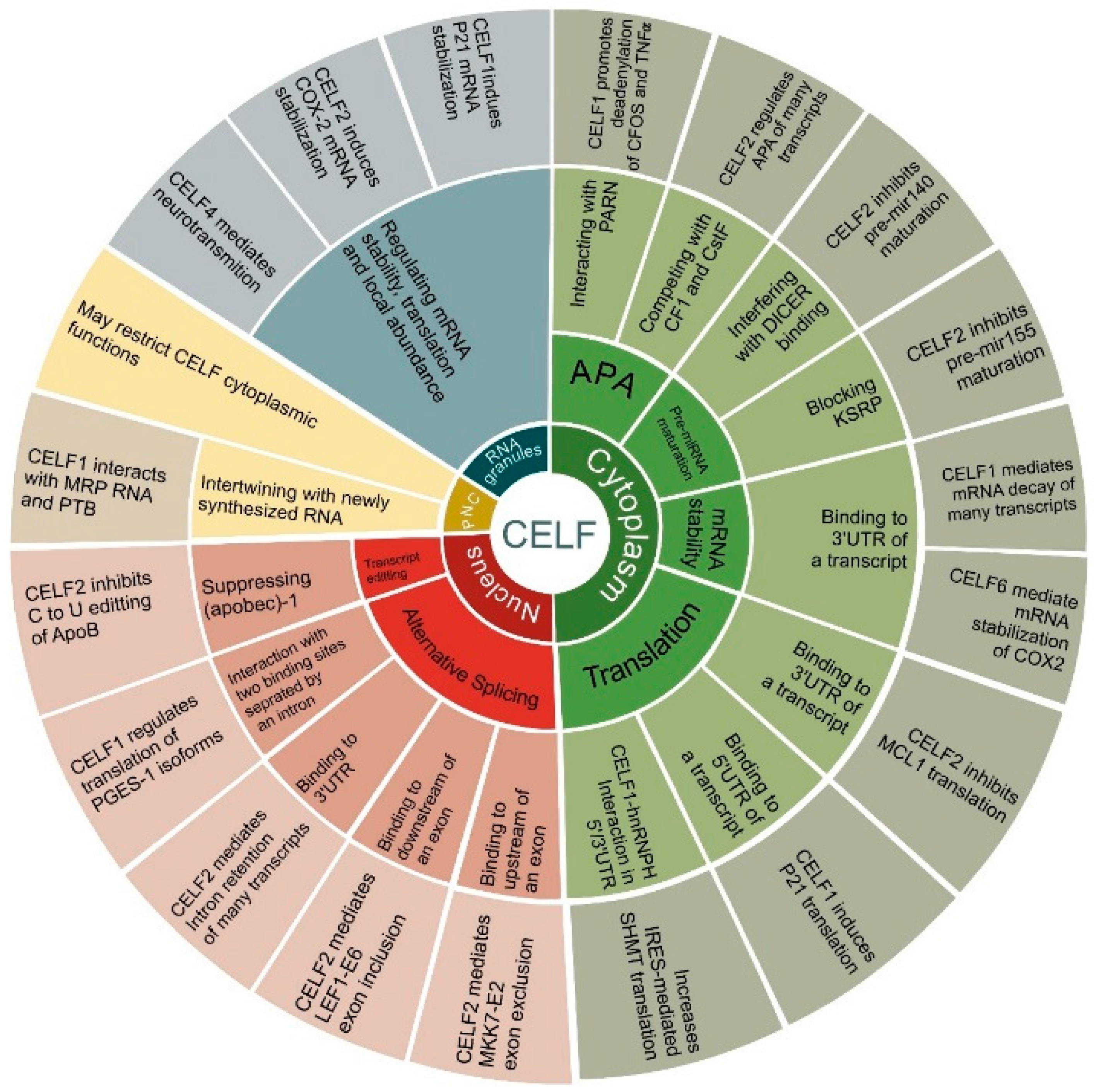
3. CELF Targets in Cancer
4. CELF Regulation by Noncoding RNAs
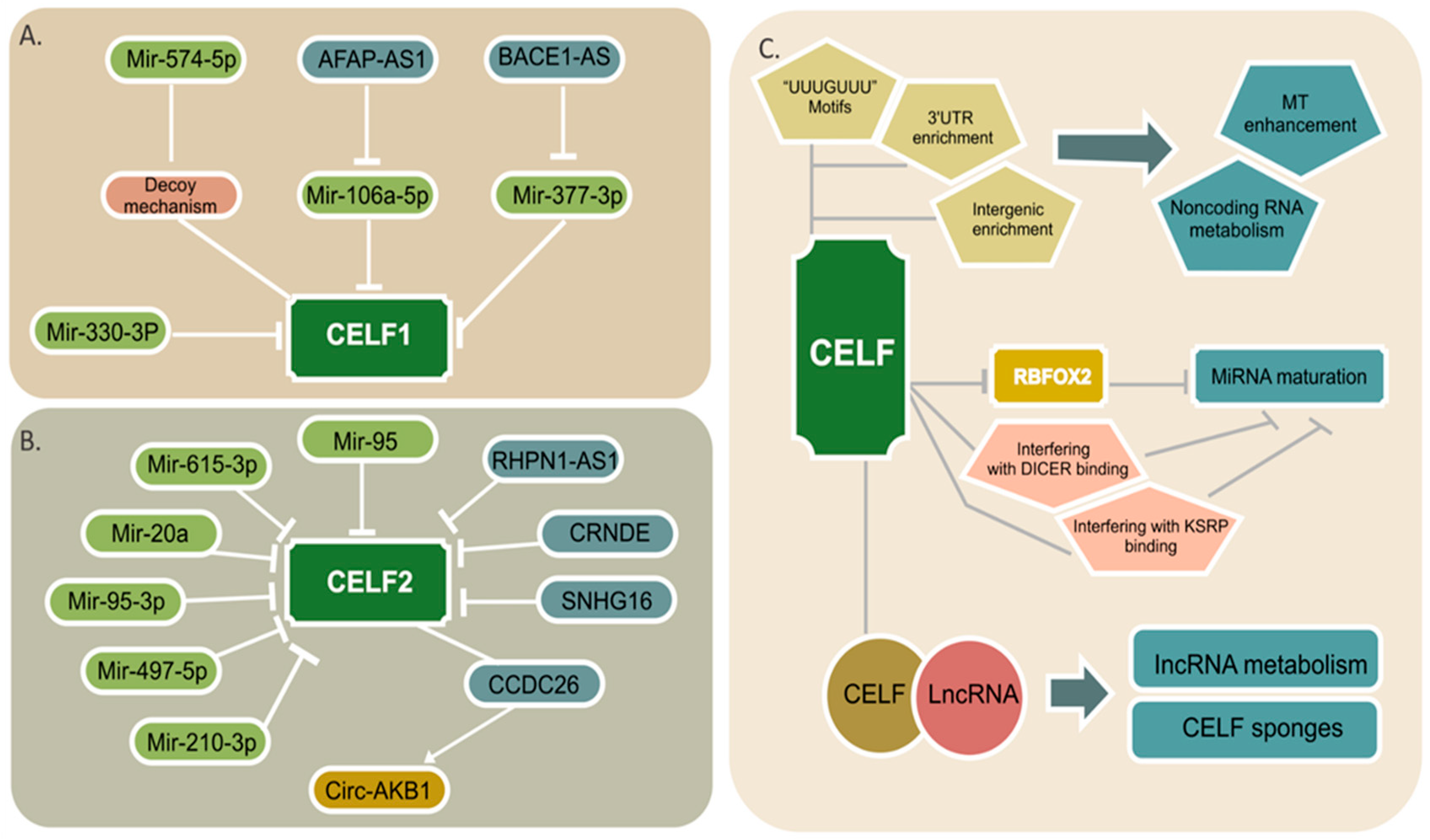
5. Noncoding RNA Regulation by CELF
This entry is adapted from the peer-reviewed paper 10.3390/ijms222011056
References
- Sternburg, E.L.; Karginov, F.V. Global Approaches in Studying RNA-Binding Protein Interaction Networks. Trends Biochem. Sci. 2020, 45, 593–603.
- Kim, S.; Kim, S.; Chang, H.R.; Kim, D.; Park, J.; Son, N.; Park, J.; Yoon, M.; Chae, G.; Kim, Y.-K.; et al. The regulatory impact of RNA-binding proteins on microRNA targeting. Nat. Commun. 2021, 12, 1–15.
- Sebestyén, E.; Singh, B.; Miñana, B.; Pagès, A.; Mateo, F.; Pujana, M.A.; Valcárcel, J.; Eyras, E. Large-scale analysis of genome and transcriptome alterations in multiple tumors unveils novel cancer-relevant splicing networks. Genome Res. 2016, 26, 732–744.
- Kechavarzi, B.; Janga, S.C. Dissecting the expression landscape of RNA-binding proteins in human cancers. Genome Biol. 2014, 15, R14.
- Dasgupta, T.; Ladd, A.N. The importance of CELF control: Molecular and biological roles of the CUG-BP, Elav-like family of RNA-binding proteins. Wiley Interdiscip. Rev. RNA 2011, 3, 104–121.
- Louis, I.V.-S.; Dickson, A.M.; Bohjanen, P.; Wilusz, C.J. CELFish ways to modulate mRNA decay. Biochim. Biophys. Acta Bioenerg. 2013, 1829, 695–707.
- Ajith, S.; Gazzara, M.R.; Cole, B.S.; Shankarling, G.; Martinez, N.; Mallory, M.J.; Lynch, K.W. Position-dependent activity of CELF2 in the regulation of splicing and implications for signal-responsive regulation in T cells. RNA Biol. 2016, 13, 569–581.
- Xia, H.; Chen, D.; Wu, Q.; Wu, G.; Zhou, Y.; Zhang, Y.; Zhang, L. CELF1 preferentially binds to exon-intron boundary and regulates alternative splicing in HeLa cells. Biochim. Biophys. Acta Bioenerg. 2017, 1860, 911–921.
- Goodall, G.J.; Wickramasinghe, V.O. RNA in cancer. Nat. Rev. Cancer 2020, 21, 22–36.
- Cursons, J.; Pillman, K.A.; Scheer, K.G.; Gregory, P.A.; Foroutan, M.; Hediyeh-Zadeh, S.; Toubia, J.; Crampin, E.J.; Goodall, G.J.; Bracken, C.P.; et al. Combinatorial Targeting by MicroRNAs Co-ordinates Post-transcriptional Control of EMT. Cell Syst. 2018, 7, 77–91.e7.
- Maloney, S.E.; Khangura, E.; Dougherty, J.D. The RNA-binding protein Celf6 is highly expressed in diencephalic nuclei and neuromodulatory cell populations of the mouse brain. Brain Struct. Funct. 2015, 221, 1809–1831.
- Rieger, M.A.; King, D.M.; Crosby, H.; Liu, Y.; Cohen, B.A.; Dougherty, J.D. CLIP and Massively Parallel Functional Analysis of CELF6 Reveal a Role in Destabilizing Synaptic Gene mRNAs through Interaction with 3′ UTR Elements. Cell Rep. 2020, 33, 108531.
- Ladd, A.; Cooper, T.A. Multiple domains control the subcellular localization and activity of ETR-3, a regulator of nuclear and cytoplasmic RNA processing events. J. Cell Sci. 2004, 117, 3519–3529.
- Itai, T.; Hamanaka, K.; Sasaki, K.; Wagner, M.; Kotzaeridou, U.; Brösse, I.; Ries, M.; Kobayashi, Y.; Tohyama, J.; Kato, M.; et al. De novo variants in CELF2 that disrupt the nuclear localization signal cause developmental and epileptic encephalopathy. Hum. Mutat. 2020, 42, 66–76.
- Salisbury, E.; Sakai, K.; Schoser, B.; Huichalaf, C.; Schneider-Gold, C.; Nguyen, H.; Wang, G.-L.; Albrecht, J.H.; Timchenko, L.T. Ectopic expression of cyclin D3 corrects differentiation of DM1 myoblasts through activation of RNA CUG-binding protein, CUGBP. Exp. Cell Res. 2008, 314, 2266–2278.
- Beisang, D.; Rattenbacher, B.; Louis, I.A.V.-S.; Bohjanen, P.R. Regulation of CUG-binding Protein 1 (CUGBP1) Binding to Target Transcripts upon T Cell Activation. J. Biol. Chem. 2012, 287, 950–960.
- Choudhary, C.; Kumar, C.; Gnad, F.; Nielsen, M.L.; Rehman, M.; Walther, T.C.; Olsen, J.V.; Mann, M. Lysine Acetylation Targets Protein Complexes and Co-Regulates Major Cellular Functions. Science 2009, 325, 834–840.
- Gnad, F.; Gunawardena, J.; Mann, M. PHOSIDA 2011: The posttranslational modification database. Nucleic Acids Res. 2010, 39, D253–D260.
- Suzuki, H.; Maegawa, S.; Nishibu, T.; Sugiyama, T.; Yasuda, K.; Inoue, K. Vegetal localization of the maternal mRNA encoding an EDEN-BP/Bruno-like protein in zebrafish. Mech. Dev. 2000, 93, 205–209.
- Choi, D.-K.; Yoo, K.-W.; Hong, S.-K.; Rhee, M.; Sakaki, Y.; Kim, C.-H. Isolation and expression of Napor/CUG-BP2 in embryo development. Biochem. Biophys. Res. Commun. 2003, 305, 448–454.
- Ramalingam, S.; Natarajan, G.; Schafer, C.; Subramaniam, D.; May, R.; Ramachandran, I.; Queimado, L.; Houchen, C.W.; Anant, S. Novel intestinal splice variants of RNA-binding protein CUGBP2: Isoform-specific effects on mitotic catastrophe. Am. J. Physiol. Liver Physiol. 2008, 294, G971–G981.
- Suzuki, H.; Takeuchi, M.; Sugiyama, A.; Alam, A.K.; Vu, L.T.; Sekiyama, Y.; Dam, H.C.; Ohki, S.-Y.; Tsukahara, T. Alternative splicing produces structural and functional changes in CUGBP. BMC Biochem. 2012, 13, 6.
- Hornbeck, P.V.; Zhang, B.; Murray, B.; Kornhauser, J.M.; Latham, V.; Skrzypek, E. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2014, 43, D512–D520.
- Fujimura, K.; Kano, F.; Murata, M. Dual localization of the RNA binding protein CUGBP-1 to stress granule and perinucleolar compartment. Exp. Cell Res. 2008, 314, 543–553.
- Wagnon, J.L.; Briese, M.; Sun, W.; Mahaffey, C.L.; Curk, T.; Rot, G.; Ule, J.; Frankel, W.N. CELF4 Regulates Translation and Local Abundance of a Vast Set of mRNAs, Including Genes Associated with Regulation of Synaptic Function. PLoS Genet. 2012, 8, e1003067.
- Cox, D.C.; Guan, X.; Xia, Z.; Cooper, T.A. Increased nuclear but not cytoplasmic activities of CELF1 protein leads to muscle wasting. Hum. Mol. Genet. 2020, 29, 1729–1744.
- Mallory, M.J.; Jackson, J.; Weber, B.; Chi, A.; Heyd, F.; Lynch, K.W. Signal- and Development-Dependent Alternative Splicing of LEF1 in T Cells Is Controlled by CELF. Mol. Cell. Biol. 2011, 31, 2184–2195.
- Nikonova, E.; Kao, S.-Y.; Ravichandran, K.; Wittner, A.; Spletter, M.L. Conserved functions of RNA-binding proteins in muscle. Int. J. Biochem. Cell Biol. 2019, 110, 29–49.
- Blech-Hermoni, Y.; Dasgupta, T.; Coram, R.J.; Ladd, A.N. Identification of Targets of CUG-BP, Elav-Like Family Member 1 (CELF1) Regulation in Embryonic Heart Muscle. PLoS ONE 2016, 11, e0149061.
- Emmerich, A.C.; Wellstein, J.; Ossipova, E.; Baumann, I.; Lengqvist, J.; Kultima, K.; Jakobsson, P.-J.; Steinhilber, D.; Saul, M.J. Proteomics-Based Characterization of miR-574-5p Decoy to CUGBP1 Suggests Specificity for mPGES-1 Regulation in Human Lung Cancer Cells. Front. Pharmacol. 2020, 11, 196.
- Gazzara, M.R.; Mallory, M.J.; Roytenberg, R.; Lindberg, J.P.; Jha, A.; Lynch, K.W.; Barash, Y. Ancient antagonism between CELF and RBFOX families tunes mRNA splicing outcomes. Genome Res. 2017, 27, 1360–1370.
- Chatrikhi, R.; Mallory, M.J.; Gazzara, M.R.; Agosto, L.M.; Zhu, W.S.; Litterman, A.J.; Ansel, K.M.; Lynch, K.W. RNA Binding Protein CELF2 Regulates Signal-Induced Alternative Polyadenylation by Competing with Enhancers of the Polyadenylation Machinery. Cell Rep. 2019, 28, 2795–2806.e3.
- Chen, Z.; Eggerman, T.L.; Patterson, A.P. ApoB mRNA editing is mediated by a coordinated modulation of multiple apoB mRNA editing enzyme components. Am. J. Physiol. Liver Physiol. 2007, 292, G53–G65.
- Mallory, M.J.; McClory, S.P.; Chatrikhi, R.; Gazzara, M.R.; Ontiveros, R.J.; Lynch, K.W. Reciprocal regulation of hnRNP C and CELF2 through translation and transcription tunes splicing activity in T cells. Nucleic Acids Res. 2020, 48, 5710–5719.
- Moraes, K.C.; Wilusz, C.J.; Wilusz, J. CUG-BP binds to RNA substrates and recruits PARN deadenylase. RNA 2006, 12, 1084–1091.
- Yoon, J.S.J.; Wu, M.K.; Zhu, T.H.; Zhao, H.; Cheung, S.T.; Chamberlain, T.; Mui, A.L.-F. Interleukin-10 control of pre-miR155 maturation involves CELF. PLoS ONE 2020, 15, e0231639.
- Dejene, E.A.; Li, Y.; Showkatian, Z.; Ling, H.; Seto, E. Regulation of poly(a)-specific ribonuclease activity by reversible lysine acetylation. J. Biol. Chem. 2020, 295, 10255–10270.
- Chaudhury, A.; Cheema, S.; Fachini, J.M.; Kongchan, N.; Lu, G.; Simon, L.M.; Wang, T.; Mao, S.; Rosen, D.G.; Ittmann, M.M.; et al. CELF1 is a central node in post-transcriptional regulatory programmes underlying EMT. Nat. Commun. 2016, 7, 13362.
- Popovitchenko, T.; Park, Y.; Page, N.F.; Luo, X.; Krsnik, Z.; Liu, Y.; Salamon, I.; Stephenson, J.D.; Kraushar, M.L.; Volk, N.L.; et al. Translational derepression of Elavl4 isoforms at their alternative 5′ UTRs determines neuronal development. Nat. Commun. 2020, 11, 1674.
- Gareau, C.; Fournier, M.-J.; Filion, C.; Coudert, L.; Martel, D.; Labelle, Y.; Mazroui, R. p21WAF1/CIP1 Upregulation through the Stress Granule-Associated Protein CUGBP1 Confers Resistance to Bortezomib-Mediated Apoptosis. PLoS ONE 2011, 6, e20254.
- Subramaniam, D.; Natarajan, G.; Ramalingam, S.; Ramachandran, I.; May, R.; Queimado, L.; Houchen, C.W.; Anant, S. Translation inhibition during cell cycle arrest and apoptosis: Mcl-1 is a novel target for RNA binding protein CUGBP. Am. J. Physiol. Liver Physiol. 2008, 294, G1025–G1032.
- Fox, J.T.; Stover, P.J. Mechanism of the Internal Ribosome Entry Site-mediated Translation of Serine Hydroxymethyltransferase 1. J. Biol. Chem. 2009, 284, 31085–31096.
- Treiber, T.; Treiber, N.; Plessmann, U.; Harlander, S.; Daiß, J.-L.; Eichner, N.; Lehmann, G.; Schall, K.; Urlaub, H.; Meister, G. A Compendium of RNA-Binding Proteins that Regulate MicroRNA Biogenesis. Mol. Cell 2017, 66, 270–284.e13.
- Amen, T.; Guihur, A.; Zelent, C.; Ursache, R.; Wilting, J.; Kaganovich, D. Resveratrol and related stilbene-derivatives induce Stress Granules with distinct clearance kinetics. Mol. Biol. Cell 2021. mbc.E21–02.
- Moraes, K.C.M.; Monteiro, C.J.; Pacheco-Soares, C. A novel function for CUGBP2 in controlling the pro-inflammatory stimulus in H9c2 cells: Subcellular trafficking of messenger molecules. Cell Biol. Int. 2013, 37, 1129–1138.
- Qian, Y.; Lv, S.; Dong, X. Detection and Significance of PNC in Breast Cancer Tissues. Clin. Lab. 2021, 67.
- Vilimas, T.; Wang, A.Q.; Patnaik, S.; Hughes, E.A.; Singleton, M.D.; Knotts, Z.; Li, D.; Frankowski, K.; Schlomer, J.J.; Guerin, T.M.; et al. Pharmacokinetic evaluation of the PNC disassembler metarrestin in wild-type and Pdx1-Cre;LSL-KrasG12D/+;Tp53R172H/+ (KPC) mice, a genetically engineered model of pancreatic cancer. Cancer Chemother. Pharmacol. 2018, 82, 1067–1080.
- Pollock, C.; Daily, K.; Nguyen, V.T.; Wang, C.; Lewandowska, M.A.; Bensaude, O.; Huang, S. Characterization of MRP RNA–protein interactions within the perinucleolar compartment. Mol. Biol. Cell 2011, 22, 858–866.
- Pereira, B.; Billaud, M.; Almeida, R. RNA-Binding Proteins in Cancer: Old Players and New Actors. Trends Cancer 2017, 3, 506–528.
- Schuschel, K.; Helwig, M.; Hüttelmaier, S.; Heckl, D.; Klusmann, J.-H.; Hoell, J.I. RNA-Binding Proteins in Acute Leukemias. Int. J. Mol. Sci. 2020, 21, 3409.
- Kozomara, A.; Griffiths-Jones, S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2013, 42, D68–D73.
- Matsuyama, H.; Suzuki, H.I. Systems and Synthetic microRNA Biology: From Biogenesis to Disease Pathogenesis. Int. J. Mol. Sci. 2019, 21, 132.
- Qin, G.; Wu, X. Hsa_circ_0032463 acts as the tumor promoter in osteosarcoma by regulating the miR-330-3p/PNN axis. Int. J. Mol. Med. 2021, 47, 1–14.
- Wang, H.; Liu, G.; Li, T.; Wang, N.; Wu, J.; Zhi, H. MiR-330-3p functions as a tumor suppressor that regulates glioma cell proliferation and migration by targeting CELF. Arch. Med Sci. 2020, 16, 1166–1175.
- Saul, M.J.; Baumann, I.; Bruno, A.; Emmerich, A.C.; Wellstein, J.; Ottinger, S.M.; Contursi, A.; Dovizio, M.; Donnini, S.; Tacconelli, S.; et al. miR-574-5p as RNA decoy for CUGBP1 stimulates human lung tumor growth by mPGES-1 induction. FASEB J. 2019, 33, 6933–6947.
- Wang, J.; Liu, L.; Sun, Y.; Xue, Y.; Qu, J.; Pan, S.; Li, H.; Qu, H.; Wang, J.; Zhang, J. miR-615-3p promotes proliferation and migration and inhibits apoptosis through its potential target CELF2 in gastric cancer. Biomed. Pharmacother. 2018, 101, 406–413.
- Wu, H.; Wei, H.; Chen, Q. Long noncoding RNA HOTTIP promotes the metastatic potential of ovarian cancer through the regulation of the miR -615-3p/ SMARCE1 pathway. Kaohsiung J. Med Sci. 2020, 36, 973–982.
- Liao, C.; Chen, W.; Wang, J. MicroRNA-20a Regulates Glioma Cell Proliferation, Invasion, and Apoptosis by Targeting CUGBP Elav-Like Family Member. World Neurosurg. 2019, 121, e519–e527.
- Fan, B.; Jiao, B.-H.; Fan, F.-S.; Lu, S.-K.; Song, J.; Guo, C.-Y.; Yang, J.-K.; Yang, L. Downregulation of miR-95-3p inhibits proliferation, and invasion promoting apoptosis of glioma cells by targeting CELF. Int. J. Oncol. 2015, 47, 1025–1033.
- Xu, H.; Wang, F.; Wang, L. Suppression of miR-106a-5p expression inhibits tumorigenesis via increasing CELF-2 expression in spinal cord glioma. Oncol. Lett. 2021, 22, 1–9.
- Ge, L.; Zhou, F.; Nie, J.; Wang, X.; Zhao, Q. Hypoxic colorectal cancer-secreted exosomes deliver miR-210-3p to normoxic tumor cells to elicit a protumoral effect. Exp. Biol. Med. 2021.
- Xiao, Z.; Chow, S.C.; Li, C.H.; Tang, S.C.; Tsui, S.K.; Lin, Z.; Chen, Y. Role of microRNA-95 in the anticancer activity of Brucein D in hepatocellular carcinoma. Eur. J. Pharmacol. 2014, 728, 141–150.
- Lee, K.H.; Lee, J.K.; Choi, D.W.; Do, I.-G.; Sohn, I.; Jang, K.-T.; Jung, S.-H.; Heo, J.S.; Choi, S.H.; Lee, K.T. Postoperative Prognosis Prediction of Pancreatic Cancer With Seven MicroRNAs. Pancreas 2015, 44, 764–768.
- Zehentmayr, F.; Hauser-Kronberger, C.; Zellinger, B.; Hlubek, F.; Schuster, C.; Bodenhofer, U.; Fastner, G.; Deutschmann, H.; Steininger, P.; Reitsamer, R.; et al. Hsa-miR-375 is a predictor of local control in early stage breast cancer. Clin. Epigenetics 2016, 8, 28.
- Zellinger, B.; Bodenhofer, U.; Engländer, I.A.; Kronberger, C.; Strasser, P.; Grambozov, B.; Fastner, G.; Stana, M.; Reitsamer, R.; Sotlar, K.; et al. Hsa-miR-375/RASD1 Signaling May Predict Local Control in Early Breast Cancer. Genes 2020, 11, 1404.
- Tang, W.; Li, G.-S.; Li, J.-D.; Pan, W.-Y.; Shi, Q.; Xiong, D.-D.; Mo, C.-H.; Zeng, J.-J.; Chen, G.; Feng, Z.-B.; et al. The role of upregulated miR-375 expression in breast cancer: An in vitro and in silico study. Pathol.-Res. Pr. 2019, 216, 152754.
- Jonsdottir, K.; Janssen, S.R.; Da Rosa, F.C.; Gudlaugsson, E.; Skaland, I.; Baak, J.P.A.; Janssen, E.A.M. Validation of Expression Patterns for Nine miRNAs in 204 Lymph-Node Negative Breast Cancers. PLoS ONE 2012, 7, e48692.
- Fang, J.; Li, Y.; Zhang, J.; Yan, M.; Li, J.; Bao, S.; Jin, T. Correlation between polymorphisms in microRNA-regulated genes and cervical cancer susceptibility in a Xinjiang Uygur population. Oncotarget 2017, 8, 31758–31764.
- Chi, Y.; Wang, J.; Yu, W.; Yang, J. Long Non-Coding RNA in the Pathogenesis of Cancers. Cells 2019, 8, 1015.
- Liu, C.; Wang, H.; Tang, L.; Huang, H.; Xu, M.; Lin, Y.; Zhou, L.; Ho, L.; Lu, J.; Ai, X. LncRNA BACE1-AS enhances the invasive and metastatic capacity of hepatocellular carcinoma cells through mediating miR-377-3p/CELF1 axis. Life Sci. 2021, 275, 119288.
- Jin, H.; Liang, G.; Yang, L.; Liu, L.; Wang, B.; Yan, F. SP1-induced AFAP1-AS1 contributes to proliferation and invasion by regulating miR-497-5p/CELF1 pathway in nasopharyngeal carcinoma. Hum. Cell 2021, 34, 491–501.
- Shi, M.; Yang, R.; Lin, J.; Wei, Q.; Chen, L.; Gong, W.; Guo, X. LncRNA-SNHG16 promotes proliferation and migration of acute myeloid leukemia cells via PTEN/PI3K/AKT axis through suppressing CELF2 protein. J. Biosci. 2021, 46, 1–13.
- Xie, S.-C.; Zhang, J.-Q.; Jiang, X.-L.; Hua, Y.-Y.; Xie, S.-W.; Qin, Y.-A.; Yang, Y.-J. LncRNA CRNDE facilitates epigenetic suppression of CELF2 and LATS2 to promote proliferation, migration and chemoresistance in hepatocellular carcinoma. Cell Death Dis. 2020, 11, 1–17.
- Zhao, Y.; Zhou, H.; Dong, W. LncRNA RHPN1-AS1 promotes the progression of nasopharyngeal carcinoma by targeting CELF2 expression. Exp. Mol. Pathol. 2021, 122, 104671.
- Li, C.; Mu, J.; Shi, Y.; Xin, H. LncRNA CCDC26 Interacts with CELF2 Protein to Enhance Myeloid Leukemia Cell Proliferation and Invasion via the circRNA_ANKIB1/miR-195-5p/PRR11 Axis. Cell Transplant. 2021, 30.
- Jacobsen, A.; Wen, J.; Marks, D.S.; Krogh, A. Signatures of RNA binding proteins globally coupled to effective microRNA target sites. Genome Res. 2010, 20, 1010–1019.
- Chen, Y.; Zubovic, L.; Yang, F.; Godin, K.; Pavelitz, T.; Castellanos, J.; Macchi, P.; Varani, G. Rbfox proteins regulate microRNA biogenesis by sequence-specific binding to their precursors and target downstream Dicer. Nucleic Acids Res. 2016, 44, 4381–4395.
- Nussbacher, J.K.; Yeo, G.W. Systematic Discovery of RNA Binding Proteins that Regulate MicroRNA Levels. Mol. Cell 2018, 69, 1005–1016.e7.
- Ruggiero, T.; Trabucchi, M.; De Santa, F.; Zupo, S.; Harfe, B.D.; McManus, M.T.; Rosenfeld, M.G.; Briata, P.; Gherzi, R. LPS induces KH-type splicing regulatory protein-dependent processing of microRNA-155 precursors in macrophages. FASEB J. 2009, 23, 2898–2908.
- Ahuja, D.; Goyal, A.; Ray, P.S. Interplay between RNA-binding protein HuR and microRNA-125b regulates p53 mRNA translation in response to genotoxic stress. RNA Biol. 2016, 13, 1152–1165.
- Liu, L.; Ouyang, M.; Rao, J.N.; Zou, T.; Xiao, L.; Chung, H.K.; Wu, J.; Donahue, J.M.; Gorospe, M.; Wang, J.-Y. Competition between RNA-binding proteins CELF1 and HuR modulates MYC translation and intestinal epithelium renewal. Mol. Biol. Cell 2015, 26, 1797–1810.
- Le Tonquèze, O.; Gschloessl, B.; Legagneux, V.; Paillard, L.; Audic, Y. Identification of CELF1 RNA targets by CLIP-seq in human HeLa cells. Genom. Data 2016, 8, 97–103.
- Ebert, M.S.; Sharp, P.A. Emerging Roles for Natural MicroRNA Sponges. Curr. Biol. 2010, 20, R858–R861.
- Dumbovic, G.; Biayna, J.; Banús, J.; Samuelsson, J.; Roth, A.; Diederichs, S.; Alonso, S.; Buschbeck, M.; Perucho, M.; Forcales, S.-V. A novel long non-coding RNA from NBL2 pericentromeric macrosatellite forms a perinucleolar aggregate structure in colon cancer. Nucleic Acids Res. 2018, 46, 5504–5524.
