Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Agriculture, Dairy & Animal Science
Microorganisms in soils and plants affect soil physical and chemical characteristics, affect soil nutrient availability and distribution and are crucial for soil and plant health, aiding in resilience to environmental stresses.
- soil and root microbiome
- field management
- 16S rRNA
- bioindicator
1. Introduction
Conventional cropping systems, which rely heavily on chemical fertilizers and control agents, have been utilized for decades, leading to a doubling of crop production and increasing food demand among the global population [1]. However, this agricultural practice has many negative effects on ecological diversity, soil structure and human health [1][2][3]. In contrast, organic cropping systems, which have been practiced for thousands of years, conserve natural sources and maintain environmental health by using organic materials and low intensive tillage, making agriculture sustainable [3]. Field management makes a significant difference and has a great impact on biological diversity, including microorganism community structures and compositions.
Previous reports have shown the effects of agricultural practices on soil and rhizosphere microbiomes. For example, soil bacterial richness and diversity was significantly increased in organic farms, compared to conventional farms [4]. Zhang et al. [5] found that the abundance of bacteria which obtain acid phosphatase activity was altered in rice when the irrigation regimes were changed. In semiarid vineyards, cover crop management not only increased soil organic carbon but also enhanced bacterial diversity compared to conventional tillage. In contrast, fungal diversity was higher under conventional tillage than under cover crop management [6]. In addition, evidence also suggested that soil fungal communities are more sensitive to crop rotation and cropping systems than bacterial communities are [7][8]. There are also reports that show the effects of field management on root endophytic bacterial microbiomes [9][10]. However, there are only a few reports that examine the impacts of field management on both soils and root microbiomes. Hartman et al. [11] reported that field management is a crucial factor in determining soil fungal structure, but in root endosphere it is the main driving force for bacteria rather than fungi. The responses of bacterial and fungal communities to agricultural practices vary based on environmental conditions and crop species, suggesting that more investigation is required to elucidate the effects of agriculture practices on soil, rhizosphere and endosphere microbiomes.
Tea (Camellia sinensis L.) is one of the most important economic crops in the world. It is a perennial crop that is grown for the harvesting of young leaves. Thus, nutrient replenishment, especially nitrogen, is crucial for tea production. However, the long-term application of chemicals is harmful to soil health and the ecosystem at large. Several studies showed that applying organic fertilizers to tea farms increases soil pH, organic carbon and total N content [12][13]. The width and length of tea leaves are also greater under organic farming systems, although yield might be reduced compared to conventional farming systems [12][14]. Moreover, leaves harvested on organic farms also enhance tea’s antioxidant properties [14][15][16]. In addition, Qiu et al. [17] examined soil microbial diversity through temperature gradient gel electrophoresis and found a lower Shannon index when applying chemical fertilizers instead of organic manure, suggesting that field management affects microbial biodiversity in tea farms. However, little is known about the microbial community structures and species that are sensitive to field management in the tea farms.
2. Identification of OTUs That Are Sensitive to Field Management and Growth Environments
The distinction of bacterial community composition among samples was demonstrated by PERMANOVA and PCoA. Next, we performed the LEfSe procedure to display how taxonomic abundance was affected by field management strategy. At all the levels, samples derived from organic farms were relatively abundant compared to those from conventional farms. One phylum (Latescibacterota), four classes, six orders and six families (LDA score > 4) were enriched in the samples from organic farms. In contrast, only one class, one order and one family were enriched in the samples from conventional farms (Figure S4).
We further identified the OTUs that were sensitive to field management strategies by performing an indicator species analysis. Among OTUs identified in the pool of soil bacteria, 11 and 26 OTUs were potential indicator species of conventional and organic farms, respectively, and five OTUs were co-occurred in both types of cropping systems, meaning that they might have been indicators of soil bacteria. Among root endophytic OTUs, 21 and 16 OTUs were identified as indicator species of root endophytes under conventional and organic cropping systems, respectively, and only two OTUs, assigned to Phenylobacterium and Thermosporothrix, were co-occurred under both cropping systems; these are the bioindicators of root bacteria. Interestingly, four OTUs were co-occurred in the roots and soils derived from conventional farms, while only one OUT, assigned to Sphingomonas, was co-occurred in the soil and root samples from organic farms, suggesting that these OTUs might be the indicator species of cropping systems in either soils or roots (Figure 1; Table S2). In summary, we identified the indicator species of different sample types under different cropping systems. Further validation will be required to establish a strong association between bacterial groups and field management.
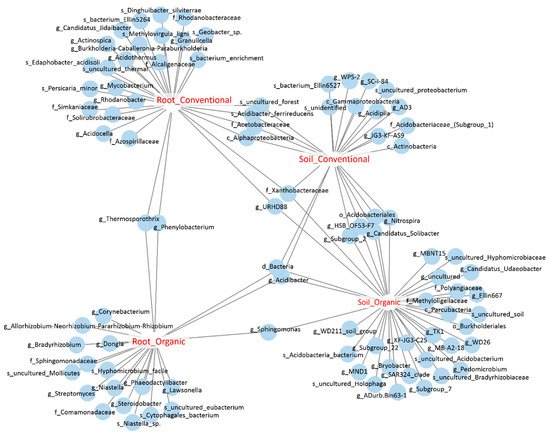
Figure 1. The bipartite network shows soil and root OTUs that were sensitive to cropping systems. Each circle represents an individual OTU that was significantly associated with the corresponding sample groups (p < 0.05).
3. Discussion
There is growing evidence to support the impacts of agriculture practices on soils, rhizosphere and root endosphere biota, including microbiota, and to suggest that the effects may be varied by crop species [4][7][9][11][18][19]. Being different from most reports that study microbial community structure and diversity in soil or root-associated compartments, our investigation examined the bacterial microbiome in both soils and tea root endosphere from three organic and conventional farms, respectively. A total of 18 soil samples and 17 root samples were harvested for analysis. Through this study, we demonstrated the impacts of field management on bacteria communities in both soils and roots.
Previous studies showed that soil organic carbon (C) and total N content was higher in organic tea farms than in conventional farms, which might lead to an increase in soil microbial biomass C, N, phosphorus content and the Shannon index [13][17]. Our work was different from previous studies that predominantly used chemicals or temperature gradient gel electrophoresis to analyze the properties of soil microbes. As such, we performed 16S rRNA amplicon-based sequencing to provide a global view of both soil and root endophytic bacterial microbiomes. First, we found that all of the α-diversity indices of soil bacterial microbiomes were higher than root endophytic bacteria. When comparing the effects of field management on α-diversity, only the evenness of soil bacteria in organic tea farms was significantly higher than in conventional farms, while no difference was observed for root endophytic bacteria. This is consistent with previous studies that suggest that species richness in soil bacterial communities is higher than in root communities; clearly, organic cropping systems enhance the α-diversity of soil bacterial communities while field management has no or marginal effects on root bacterial communities [4][9][10][11].
Root endophytes are transmitted from seeds or recruited from soils [20][21]. The strength of physical barrier in roots is one of the selection forces for soil bacteria to pass. Only a limited number of bacteria species can successfully colonize root cells [22]. Thus, it is not surprising to see a lower α-diversity in root bacterial populations than in soil, as well as a difference in soil and root bacterial community structure. When examining the variation of bacterial community composition using PCoA and PERMANOVA, field management, sample type and the interaction between the two variants significantly affected bacterial community structures. The difference between soil bacterial communities under different cropping systems might affect rhizosphere bacterial composition, which is the pool of root endophytes. At the phylum level, Proteobacteria, Actinobacteriota and Chloroflexi were the three major phyla in roots, and they occupied more than 80% abundance, while, in soils, Proteobacteria, Acidobacteriota and Chloroflexi were the three dominant phyla. Although Proteobacteria was abundant both in soil and in root communities, its relative abundance in soils was significantly lower than in roots. In addition, Actinobacteriota and Bacteroidota were also enriched in roots rather than in soils, while Acidobacteriota, Gemmatinonadota and Nitrospirota were more abundant in soils than in roots; this finding is partly consistent with a study on winter wheat [11]. Next, we investigated the effects of field management on soil and root bacterial communities, respectively. We observed the enrichment of Verrucomicrobia, Plantomycetota and Gemmatinonadota in soil bacterial communities derived from organic farms. In contrast, Verrucomicrobia and Acidobacteriota were abundant in roots under conventional cropping systems and Bacterioidota was enriched in roots under organic cropping systems. Verrucomicrobia is a ubiquitous bacterial phylum in soils and other studies even identified Verrucomicrobia communities in the rhizosphere and root endosphere [23][24], suggesting that this phylum might interact with plants. A large-scale study of tallgrass prairie soils showed that the abundance of Verrucomicrobia is affected by climate conditions. In addition, it is positively correlated with carbon metabolism function but negatively correlated with nitrogen metabolism [25]. Another study showed that Verrucomicrobia communities decrease when soil fertility increases [26], supporting the notion that this phylum may be involved in complex carbohydrate degradation and could have a high affinity for nutrients. Some studies showed that the relative abundance of Verrucomicrobia was not affected by cropping systems or even lower in organic-farmed soils [27][28], which is different to what we observed. Comparisons between climate conditions regarding this and previous studies, the latter of which were conducted in temperate climates, might suggest that the difference in results could be attributed to the carbon sources, nutrient availability and consumption. Moreover, the difference in nutrient demand and utility between vegetable crops and tea might also lead to a different response to a novel farming system, thus affecting the microbial community structure. At the genus level, Acidothermus and Rhodanobacter were enriched in roots from conventional fields. Previous studies showed that Acidothermus and Rhodanobacter preferred saline and acidic environments, respectively [23][29]. It is known that the long-term application of chemical fertilizers affects soil pH and salt content, and has a great impact on microbial community composition in the rhizosphere [30][31]. Further studies will be required to examine the seasonal change in soil fertility, plant nutrition status and microbiome to reveal the association between microbiomes and other environmental factors.
We further identified the indicator species in soils and roots that were sensitive to field management. In soils, most OTUs that were enriched in conventional farms with more than 1% relative abundance belonged to Gemmaproteobacteria, while three out of four organic farm-enriched OTUs, with more than 1% abundance, belonged to Acidobacteriae. Previous studies showed that the long-term application of chemical fertilizers increases the abundance of Gemmaproteobacteria and Acidobacteriae in the 0–10 cm soil compared to no fertilizer control, and applying organic-inorganic mixed fertilizers further boosts their abundance [32][33]. Enebe and Babalola [34] also observed an increase in Gemmaproteobacteria when applying higher amounts of inorganic fertilizer. These pieces of evidence support the notion that an abundance of Gemmaproteobacteria is highly associated with chemical fertilizers. Similarly, we also found that most OTUs enriched in roots from conventional farms with more than 1% abundance belonged to Gemmaproteobacteria, while others belonged to Alphaproteobacteria, Actinobacteria and other classes. Interestingly, in the roots from organic farms, we found an increased abundance of Bradyrhizobium, Streptomyces, Burkholderia-Caballeronia-Paraburkholderia, and Sphingomonas, all of which are genera enriched by well-known plant growth-promoting bacteria [35][36][37][38]. This result is consistent with a recent report by Reid et al. [31] showing a reduction in the population of plant growth-promoting bacteria in the rhizosphere after applying chemical fertilizers. Through both previous studies and this present study, we can see the benefit of organic cropping systems on microbial composition, which might increase crop growth and resilience to environmental stresses.
In order to understand the function of bacterial communities, functional analysis was performed, and 28 and 38 level-three KEGG categories in soil and root microbiota, which were significantly affected by field management, were identified. In soils, it is worth noting that a few pathways related to xenobiotics degradation, including “aminobenzoate degradation”, “benzoate degradation” and “naphthalene degradation” were more abundant in organic farms than in conventional farms. Xenobiotics are chemicals that accumulate in the environment that are harmful to human and environmental health. The biodegradation of xenobiotics is one effective way of removing the toxins [39]. The increase in xenobiotics’ degradation capability in organic farms suggests that soil bacteria might be capable of degrading xenobiotics and removing toxic chemicals. Further analysis is required to validate the presence of xenobiotics degradation-related genes and enzyme activity in organic soils. In roots, we noticed that two KEGG functional groups, “ABC transporters” and “transporters” were enriched in samples from organic fields, which are vital for bacterial growth and survival [40][41]. A study in Allium root bacterial endophytes showed that “ABC transporters”, “transporters” and “secretion system” were the richest pathway [42]. Here we further found that these functions are more abundant in roots under organic cropping systems. More research is needed to ascertain whether these functions benefit the nutrient uptake and stress tolerance of plants.
This entry is adapted from the peer-reviewed paper 10.3390/applmicrobiol1020025
References
- Tilman, D. Global environmental impacts of agricultural expansion: The need for sustainable and efficient practices. Proc. Natl. Acad. Sci. USA 1999, 96, 5995–6000.
- Pimentel, D.; Harvey, C.; Resosudarmo, P.; Sinclair, K.; Kurz, D.; McNair, M.; Crist, S.; Shpritz, L.; Fitton, L.; Saffouri, R.; et al. Environmental and Economic Costs of Soil Erosion and Conservation Benefits. Science 1995, 267, 1117–1123.
- Pimentel, D.; Hepperly, P.; Hanson, J.; Douds, D.; Seidel, R. Environmental, Energetic, and Economic Comparisons of Organic and Conventional Farming Systems. Bioscience 2005, 55, 573–582.
- Ishaq, S.; Johnson, S.P.; Miller, Z.J.; Lehnhoff, E.A.; Olivo, S.; Yeoman, C.J.; Menalled, F.D. Impact of Cropping Systems, Soil Inoculum, and Plant Species Identity on Soil Bacterial Community Structure. Microb. Ecol. 2016, 73, 417–434.
- Zhang, Y.; Wang, X.; Xu, F.; Song, T.; Du, H.; Gui, Y.; Xu, M.; Cao, Y.; Dang, X.; Rensing, C.; et al. Combining Irrigation Scheme and Phosphorous Application Levels for Grain Yield and Their Impacts on Rhizosphere Microbial Communities of Two Rice Varieties in a Field Trial. J. Agric. Food Chem. 2019, 67, 10577–10586.
- Novara, A.; Catania, V.; Tolone, M.; Gristina, L.; Laudicina, V.A.; Quatrini, P. Cover Crop Impact on Soil Organic Carbon, Nitrogen Dynamics and Microbial Diversity in a Mediterranean Semiarid Vineyard. Sustainability 2020, 12, 3256.
- Benitez, M.-S.; Osborne, S.L.; Lehman, R.M. Previous Crop and Rotation History Effects On Maize Seedling Health and Associated Rhizosphere Microbiome. Sci. Rep. 2017, 7, 15709.
- Morrison-Whittle, P.; Lee, S.A.; Goddard, M.R. Fungal Communities Are Differentially Affected by Conventional and Biodynamic Agricultural Management Approaches in Vineyard Ecosystems. Agric. Ecosyst. Environ. 2017, 246, 306–313.
- Wemheuer, F.; Kaiser, K.; Karlovsky, P.; Daniel, R.; Vidal, S.; Wemheuer, B. Bacterial Endophyte Communities of Three Agricultural Important Grass Species Differ in Their Response Towards Management Regimes. Sci. Rep. 2017, 7, 40914.
- Lin, G.-Y.; Lin, C.-Y.; Chang, S.-J.; Lin, W.-Y. The Dynamics of Endophytic Bacterial Community Structure in Rice Roots under Different Field Management Systems. Agronomy 2020, 10, 1623.
- Hartman, K.; Van Der Heijden, M.G.A.; Wittwer, R.A.; Banerjee, S.; Walser, J.-C.; Schlaeppi, K. Cropping Practices Manipulate Abundance Patterns of Root and Soil Microbiome Members Paving The Way to Smart Farming. Microbiome 2018, 6, 14.
- Chong, K.P.; Ho, T.Y.; Jalloh, M.B. Soil Nitrogen Phosphorus and Tea Leaf Growth in Organic and Conventional Farming of Selected Fields at Sabah Tea Plantation Slope. J. Sustain. Dev. 2009, 1, 117–122.
- Han, W.-Y.; Xu, J.-M.; Wei, K.; Shi, R.-Z.; Ma, L.-F. Soil Carbon Sequestration, Plant Nutrients and Biological Activities Affected by Organic Farming System in Tea (Camellia sinensis (L.) O. Kuntze) fields. Soil Sci. Plant Nutr. 2013, 59, 727–739.
- Das, S.; Borua, P.K.; Bhagat, R.M. Soil Nitrogen and Tea Leaf Properties in Organic and Conventional Farming Systems Under Humid Sub-Tropical Conditions. Org. Agric. 2016, 6, 119–132.
- Ghosh, B.C.; Palit, S.; Gupta, S.D.; Swain, D.K. Studies on Tea Quality Grown Through Conventional and Organic Management Practices: Its Impact on Antioxidant and Antidiarrhoeal Activity. Trans. ASABE 2008, 51, 2227–2238.
- Bagchi, A.; Ch, B.; Ghosh, R.; Swain, D.K.; Bera, N. Organic Farming Practice for Quality Improvement of Tea and Its Anti Parkinsonism Effect on Health Defense. J. Phys. Chem. Biophys. 2015, 5, 178.
- Qiu, S.-L.; Wang, L.-M.; Huang, D.-F.; Lin, X.-J. Effects of Fertilization Regimes on Tea Yields, Soil Fertility, and Soil Microbial Diversity. Chil. J. Agric. Res. 2014, 74, 333–339.
- Rodríguez-Blanco, A.; Sicardi, M.; Frioni, L. Plant Genotype and Nitrogen Fertilization Effects on Abundance and Diversity of Diazotrophic Bacteria Associated with Maize (Zea mays L.). Biol. Fertil. Soils 2015, 51, 391–402.
- Chávez-Romero, Y.; Navarro-Noya, Y.E.; Reynoso-Martínez, S.C.; Sarria-Guzmán, Y.; Govaerts, B.; Verhulst, N.; Dendooven, L.; Luna-Guido, M. 16S Metagenomics Reveals Changes in The Soil Bacterial Community Driven by Soil Organic C, N-Fertilizer and Tillage-Crop Residue Management. Soil Tillage Res. 2016, 159, 1–8.
- Hallmann, J.; Quadt-Hallmann, A.; Mahaffee, W.F.; Kloepper, J.W. Bacterial Endophytes in Agricultural Crops. Can. J. Microbiol. 1997, 43, 895–914.
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; Van Themaat, E.V.L.; Schulze-Lefert, P. Structure and Functions of the Bacterial Microbiota of Plants. Annu. Rev. Plant Biol. 2013, 64, 807–838.
- Fitzpatrick, C.R.; Copeland, J.; Wang, P.W.; Guttman, D.S.; Kotanen, P.M.; Johnson, M.T.J. Assembly and Ecological Function of The Root Microbiome Across Angiosperm Plant Species. Proc. Natl. Acad. Sci. USA 2018, 115, E1157–E1165.
- Aguirre-Von-Wobeser, E.; Rocha-Estrada, J.; Shapiro, L.R.; De La Torre, M. Enrichment of Verrucomicrobia, Actinobacteria and Burkholderiales drives selection of bacterial community from soil by maize roots in a traditional milpa agroecosystem. PLoS ONE 2018, 13, e0208852.
- Bünger, W.; Jiang, X.; Müller, J.; Hurek, T.; Reinhold-Hurek, B. Novel Cultivated Endophytic Verrucomicrobia Reveal Deep-Rooting Traits of Bacteria to Associate with Plants. Sci. Rep. 2020, 10, 8692.
- Fierer, N.; Ladau, J.; Clemente, J.C.; Leff, J.W.; Owens, S.M.; Pollard, K.S.; Knight, R.; Gilbert, J.A.; McCulley, R.L. Reconstructing the Microbial Diversity and Function of Pre-Agricultural Tallgrass Prairie Soils in the United States. Science 2013, 342, 621–624.
- Navarrete, A.; Soares, T.; Rossetto, R.; Van Veen, J.A.; Tsai, S.M.; Kuramae, E.E. Verrucomicrobial community structure and abundance as indicators for changes in chemical factors linked to soil fertility. Antonie Leeuwenhoek 2015, 108, 741–752.
- Bonanomi, G.; De Filippis, F.; Cesarano, G.; La Storia, A.; Ercolini, D.; Scala, F. Organic farming induces changes in soil microbiota that affect agro-ecosystem functions. Soil Biol. Biochem. 2016, 103, 327–336.
- Lupatini, M.; Korthals, G.W.; De Hollander, M.; Janssens, T.K.; Kuramae, E.E. Soil Microbiome Is More Heterogeneous in Organic Than in Conventional Farming System. Front. Microbiol. 2017, 7, 2064.
- Green, S.; Prakash, O.; Jasrotia, P.; Overholt, W.A.; Cardenas, E.; Hubbard, D.; Tiedje, J.M.; Watson, D.B.; Schadt, C.W.; Brooks, S.; et al. Denitrifying Bacteria from the Genus Rhodanobacter Dominate Bacterial Communities in the Highly Contaminated Subsurface of a Nuclear Legacy Waste Site. Appl. Environ. Microbiol. 2012, 78, 1039–1047.
- Citak, S.; Sonmez, S. Effects of chemical fertilizer and different organic manures application on soil pH, EC and organic matter content. J. Food Agri. Environ. 2011, 9, 739–741.
- Reid, T.E.; Kavamura, V.N.; Abadie, M.; Torres-Ballesteros, A.; Pawlett, M.; Clark, I.M.; Harris, J.; Mauchline, T.H. Inorganic Chemical Fertilizer Application to Wheat Reduces the Abundance of Putative Plant Growth-Promoting Rhizobacteria. Front. Microbiol. 2021, 12, 642587.
- Wang, L.; Li, J.; Yang, F.; E, Y.; Raza, W.; Huang, Q.; Shen, Q. Application of Bioorganic Fertilizer Significantly Increased Apple Yields and Shaped Bacterial Community Structure in Orchard Soil. Microb. Ecol. 2016, 73, 404–416.
- Ma, B.; Lv, X.; Cai, Y.; Chang, S.X.; Dyck, M. Liming does not counteract the influence of long-term fertilization on soil bacterial community structure and its co-occurrence pattern. Soil Biol. Biochem. 2018, 123, 45–53.
- Enebe, M.C.; Babalola, O.O. Effects of inorganic and organic treatments on the microbial community of maize rhizosphere by a shotgun metagenomics approach. Ann. Microbiol. 2020, 70, 49.
- Pan, F.; Meng, Q.; Wang, Q.; Luo, S.; Chen, B.; Khan, K.Y.; Yang, X.; Feng, Y. Endophytic bacterium Sphingomonas SaMR12 promotes cadmium accumulation by increasing glutathione biosynthesis in Sedum alfredii Hance. Chemosphere 2016, 154, 358–366.
- Backer, R.; Rokem, J.S.; Ilangumaran, G.; Lamont, J.; Praslickova, D.; Ricci, E.; Subramanian, S.; Smith, D.L. Plant Growth-Promoting Rhizobacteria: Context, Mechanisms of Action, and Roadmap to Commercialization of Biostimulants for Sustainable Agriculture. Front. Plant Sci. 2018, 9, 1473.
- Luo, Y.; Wang, F.; Huang, Y.; Zhou, M.; Gao, J.; Yan, T.; Sheng, H.; An, L. Sphingomonas sp. Cra20 Increases Plant Growth Rate and Alters Rhizosphere Microbial Community Structure of Arabidopsis thaliana Under Drought Stress. Front. Microbiol. 2019, 10, 1221.
- Amaresan, N.; Kumar, K.; Naik, J.H.; Bapatla, K.G.; Mishra, R.K. Streptomyces in plant growth promotion: Mechanisms and role. In New and Future Developments in Microbial Biotechnology and Bioengineering; Singh, B.P., Gupta, V.K., Passari, A.K., Eds.; Elsevier: Amsterdam, The Netherlands, 2018.
- Mishra, S.; Lin, Z.; Pang, S.; Zhang, W.; Bhatt, P.; Chen, S. Recent Advanced Technologies for the Characterization of Xenobiotic-Degrading Microorganisms and Microbial Communities. Front. Bioeng. Biotechnol. 2021, 9, 632059.
- Garai, P.; Chandra, K.; Chakravortty, D. Bacterial peptide transporters: Messengers of nutrition to virulence. Virulence 2016, 8, 297–309.
- Piepenbreier, H.; Fritz, G.; Gebhard, S. Transporters as information processors in bacterial signalling pathways. Mol. Microbiol. 2017, 104, 1–15.
- Huang, Y. Illumina-based Analysis of Endophytic Bacterial Diversity of Four Allium Species. Sci. Rep. 2019, 9, 15271.
This entry is offline, you can click here to edit this entry!
