Heat-shock proteins (HSPs) are molecular chaperones participating primarily in protein folding preventing protein degradation and subsequent cellular distress.
1. Introduction
Heat-shock proteins (HSPs) are molecular chaperones participating primarily in protein folding preventing protein degradation and subsequent cellular distress [1]. HSPs are regulated through heat-shock factor 1(HSF-1) [2]. In the steady state HSF-1 is bound to HSP90 or HSP70 [3,4]. Upon stressful signals HSF-1 dissociates from HSPs and translocates into the nucleus where it stimulates HSP expression [1]. HSPs can be exposed to the immune system through tissue necrosis and the resultant cellular debris, via organized release of exosomes/endosomes, or through their presence on the cellular membrane [5,6,7]. Their evolutionary conservation can elicit interspecies immune recognition [8]. The resulting immune response can be either immunoregulatory or immunostimulatory [9,10]. Furthermore specific HSP domains as well as certain HSP isoforms and their client proteins induce a differential autoimmune response.
2. Structure and Subcellular Localization of the Small HSP Family
The small HSP (sHSP) gene family has 11 family members [
11], which are located in the nucleus, cytoplasm, extracellular space, and the cytoskeleton where they can modulate its structure [
12,
13,
14]
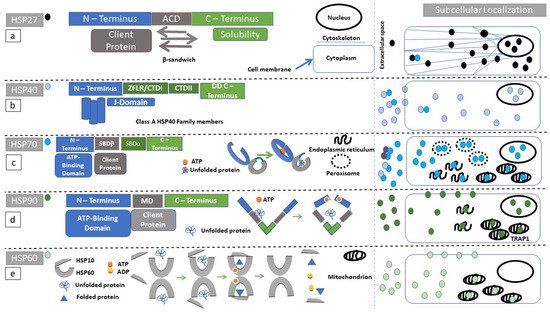
Small HSPs have a central alpha crystallin domain (ACD) bounded by N-terminal and C-terminal domains (
Figure 1a) [
15,
16,
17]. The ACD entails many antiparallel β-sheets which form its final β-sandwich conformation [
15]. The N-terminal domain contains serine residues which can be phosphorylated by intracellular kinases. For example, MAPK-activated protein kinase 5 (MK5) can interact with HSP27 in vivo and influence F-actin-dependent cytoskeletal organization [
18]. Binding of denatured proteins (client proteins) to sHSPs is characterized by diversity in terms of their docking sites. The N-terminal domain as well as the ACD can serve as client protein-binding sites [
15,
19].
Figure 1. Structure and function of heat shock proteins (HSPs): Diagrammatic representation of the domain structure and subcellular localization of HSPs under discussion. Of note is the fact that heat-shock proteins can form complexes with other molecular chaperones. These chaperone complexes may exert a different action than the uncomplexed HSPs. (a) HSP27 (black circle) secondary structure, consists of an N-terminal (blue rectangle) substrate-binding region, followed by an alpha crystallin domain (ACD, gray rectangle) ending in the C-terminus (green rectangle). ACD has a β-sandwich conformation. Client proteins dock to ACD. The C-terminus is highly variable among protein members and facilitates HSP27 oligomerization. (b) Class A HSP40 (blue circle) protein family secondary structure consists of an N-terminal (blue rectangle) substrate-binding region, followed by a zinc finger-like region (ZFLR), C-terminal domains I and II (CTDI and II, green rectangles in c-terminal region), and ending in a dimerization domain (DD). The J-domain localizes within N-terminal region. Class B preserves the N-terminal localization of the J-domain but the C-terminus can acquire a more diverse structure. In class C, the J-domain can be localized anywhere within the amino-acid sequence. (c) HSP70 (turquoise circle) secondary structure consists of an N-terminal domain (blue rectangle), followed by a substrate-binding domain (SBDβ, gray rectangle), a substrate-binding domain α-helical (SBDα, gray rectangle), and ending in the C-terminus (green rectangle). The reaction cycle involves ATP docking within N-terminal domain since ATP hydrolysis powers the structural opening of the substrate cleft within the SBDβ (gray arc). (d) The HSP90 (dark green circle) secondary structure consists of an N-terminus (blue rectangle), followed by a middle domain (MD, gray rectangle), ending in a c-terminus (green rectangle). HSP90 homodimerizes with the use of its c-terminal region. Unfolded proteins are docking in the MD. ATP hydrolysis is required for substrate processing. (e) The HSP60 (light green circle) reaction cycle. Unfolded substrates enter the HSP60 processing cleft. HSP10 acts as a lid, and ATP-hydrolysis is necessary for substrate folding.
Each one of the sHSPs plays a pivotal role in stabilizing denatured native proteins. They lack, however, the ability to refold destabilized proteins [
20], thus sHSP interaction with larger HSPs such as HSP40 or HSP70 is necessary [
15]. Larger HSPs, in contrast with sHSPs, have an ATPase function which provides the energy needed to refold the client protein [
21]. Normally sHSP molecules are in a polymeric/oligomeric state equilibrium. The presence of noxious stimuli favors their oligomerization. N- and C-termini confer to sHSPs solubility facilitating their oligomerization (
Figure 1a) [
15,
22]. sHSP oligomers can be engaged within protein aggregates in order to facilitate protein folding [
23,
24].
This entry is adapted from the peer-reviewed paper 10.3390/cells10102626