Almost 25 years have passed since a mutation of a formin gene, DIAPH1, was identified as being responsible for a human inherited disorder: a form of sensorineural hearing loss. Since then, our knowledge of the links between formins and disease has deepened considerably. Mutations of DIAPH1 and six other formin genes (DAAM2, DIAPH2, DIAPH3, FMN2, INF2 and FHOD3) have been identified as the genetic cause of a variety of inherited human disorders, including intellectual disability, renal disease, peripheral neuropathy, thrombocytopenia, primary ovarian insufficiency, hearing loss and cardiomyopathy.
- formins
- genetic disease
- gene variants
- developmental defects
- actin
1. Introduction
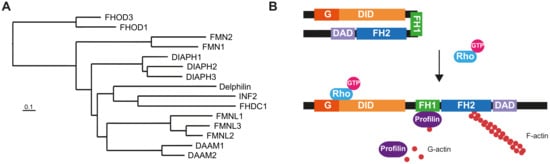
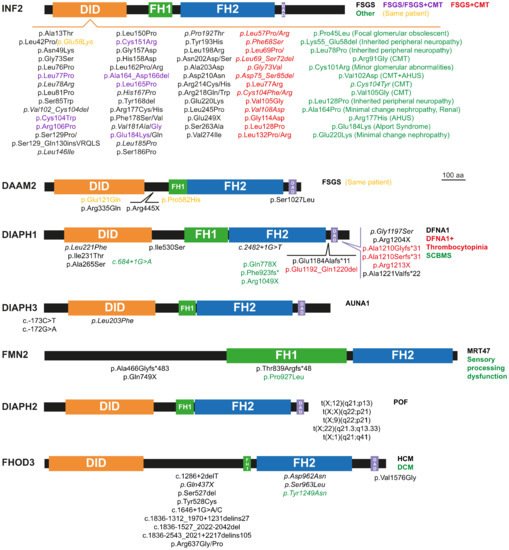
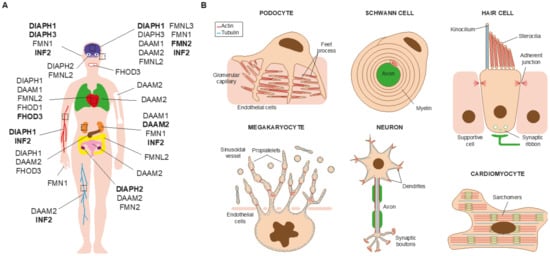
2. Monogenic Disorders Caused by Formin Mutation
2.1. Nephrotic Syndrome and Charcot-Marie-Tooth Disease
2.2. Hearing Loss
2.3. Thrombocytopenia
2.4. Microcephaly and Intellectual Disability
2.5. Primary Ovarian Insufficiency
2.6. Cardiomyopathy
This entry is adapted from the peer-reviewed paper 10.3390/cells10102554
References
- Schönichen, A.; Geyer, M. Fifteen Formins for an Actin Filament: A Molecular View on the Regulation of Human Formins. Biochim. Biophys. Acta BBA Mol. Cell Res. 2010, 1803, 152–163.
- Chesarone, M.A.; DuPage, A.G.; Goode, B.L. Unleashing Formins to Remodel the Actin and Microtubule Cytoskeletons. Nat. Rev. Mol. Cell Biol. 2010, 11, 62–74.
- Goode, B.L.; Eck, M.J. Mechanism and Function of Formins in the Control of Actin Assembly. Annu. Rev. Biochem. 2007, 76, 593–627.
- Bartolini, F.; Gundersen, G.G. Formins and Microtubules. Biochim. Biophys. Acta BBA Mol. Cell Res. 2010, 1803, 164–173.
- Fernandez-Barrera, J.; Alonso, M.A. Coordination of Microtubule Acetylation and the Actin Cytoskeleton by Formins. Cell. Mol. Life Sci. 2018, 75, 3181–3191.
- Lynch, E.D.; Lee, M.K.; Morrow, J.E.; Welcsh, P.L.; León, P.E.; King, M.-C. Nonsyndromic Deafness DFNA1 Associated with Mutation of a Human Homolog of the Drosophila Gene Diaphanous. Science 1997, 278, 1315–1318.
- Amberger, J.S.; Bocchini, C.A.; Schiettecatte, F.; Scott, A.F.; Hamosh, A. OMIM.Org: Online Mendelian Inheritance in Man (OMIM®), an Online Catalog of Human Genes and Genetic Disorders. Nucleic Acids Res. 2015, 43, D789–D798.
- Hamosh, A.; Amberger, J.S.; Bocchini, C.; Scott, A.F.; Rasmussen, S.A. Online Mendelian Inheritance in Man (OMIM®): Victor McKusick’s Magnum Opus. Am. J. Med. Genet. Part A 2021.
- Iwasa, Y.; Nishio, S.; Usami, S. Comprehensive Genetic Analysis of Japanese Autosomal Dominant Sensorineural Hearing Loss Patients. PLoS ONE 2016, 11, e0166781.
- Kaustio, M.; Nayebzadeh, N.; Hinttala, R.; Tapiainen, T.; Åström, P.; Mamia, K.; Pernaa, N.; Lehtonen, J.; Glumoff, V.; Rahikkala, E.; et al. Loss of DIAPH1 Causes SCBMS, Combined Immunodeficiency, and Mitochondrial Dysfunction. J. Allergy Clin. Immunol. 2021, 148, 599–611.
- Brozkova, D.S.; Marková, S.P.; Mészárosová, A.U.; Jenčík, J.; Čejnová, V.; Čada, Z.; Laštůvková, J.; Rašková, D.; Seeman, P. Spectrum and Frequencies of Non GJB2 Gene Mutations in Czech Patients with Early Non-Syndromic Hearing Loss Detected by Gene Panel NGS and Whole-Exome Sequencing. Clin. Genet. 2020, 98, 548–554.
- Kim, B.J.; Ueyama, T.; Miyoshi, T.; Lee, S.; Han, J.H.; Park, H.-R.; Kim, A.R.; Oh, J.; Kim, M.Y.; Kang, Y.S.; et al. Differential Disruption of Autoinhibition and Defect in Assembly of Cytoskeleton during Cell Division Decide the Fate of Human DIAPH1-Related Cytoskeletopathy. J. Med. Genet. 2019, 56, 818–827.
- Kang, T.-H.; Baek, J.-I.; Sagong, B.; Park, H.-J.; Park, C.I.; Lee, K.-Y.; Kim, U.-K. A Novel Missense Variant in the DIAPH1 Gene in a Korean Family with Autosomal Dominant Nonsyndromic Hearing Loss. Genes Genet. Syst. 2016, 91, 289–292.
- Ercan-Sencicek, A.G.; Jambi, S.; Franjic, D.; Nishimura, S.; Li, M.; El-Fishawy, P.; Morgan, T.M.; Sanders, S.J.; Bilguvar, K.; Suri, M.; et al. Homozygous Loss of DIAPH1 Is a Novel Cause of Microcephaly in Humans. Eur. J. Hum. Genet. 2015, 23, 165–172.
- Sommen, M.; Schrauwen, I.; Vandeweyer, G.; Boeckx, N.; Corneveaux, J.J.; van den Ende, J.; Boudewyns, A.; De Leenheer, E.; Janssens, S.; Claes, K.; et al. DNA Diagnostics of Hereditary Hearing Loss: A Targeted Resequencing Approach Combined with a Mutation Classification System. Hum. Mutat. 2016, 37, 812–819.
- Al-Maawali, A.; Barry, B.J.; Rajab, A.; El-Quessny, M.; Seman, A.; Coury, S.N.; Barkovich, A.J.; Yang, E.; Walsh, C.A.; Mochida, G.H.; et al. Novel Loss-of-Function Variants in DIAPH1 Associated with Syndromic Microcephaly, Blindness, and Early Onset Seizures. Am. J. Med. Genet. Part A 2016, 170, 435–440.
- Yavarna, T.; Al-Dewik, N.; Al-Mureikhi, M.; Ali, R.; Al-Mesaifri, F.; Mahmoud, L.; Shahbeck, N.; Lakhani, S.; AlMulla, M.; Nawaz, Z.; et al. High Diagnostic Yield of Clinical Exome Sequencing in Middle Eastern Patients with Mendelian Disorders. Hum. Genet. 2015, 134, 967–980.
- Wu, K.; Wang, H.; Guan, J.; Lan, L.; Zhao, C.; Zhang, M.; Wang, D.; Wang, Q. A Novel Variant in Diaphanous Homolog 1 (DIAPH1) as the Cause of Auditory Neuropathy in a Chinese Family. Int. J. Pediatr. Otorhinolaryngol. 2020, 133, 109947.
- Westbury, S.K.; Downes, K.; Burney, C.; Lozano, M.L.; Obaji, S.G.; Toh, C.H.; Sevivas, T.; Morgan, N.V.; Erber, W.N.; Kempster, C.; et al. Phenotype Description and Response to Thrombopoietin Receptor Agonist in DIAPH1-Related Disorder. Blood Adv. 2018, 2, 2341–2346.
- Shearer, A.E.; Black-Ziegelbein, E.A.; Hildebrand, M.S.; Eppsteiner, R.W.; Ravi, H.; Joshi, S.; Guiffre, A.C.; Sloan, C.M.; Happe, S.; Howard, S.D.; et al. Advancing Genetic Testing for Deafness with Genomic Technology. J. Med. Genet. 2013, 50, 627–634.
- Ueyama, T.; Ninoyu, Y.; Nishio, S.; Miyoshi, T.; Torii, H.; Nishimura, K.; Sugahara, K.; Sakata, H.; Thumkeo, D.; Sakaguchi, H.; et al. Constitutive Activation of DIA1 (DIAPH1) via C-terminal Truncation Causes Human Sensorineural Hearing Loss. EMBO Mol. Med. 2016, 8, 1310–1324.
- Neuhaus, C.; Lang-Roth, R.; Zimmermann, U.; Heller, R.; Eisenberger, T.; Weikert, M.; Markus, S.; Knipper, M.; Bolz, H.J. Extension of the Clinical and Molecular Phenotype of DIAPH1-Associated Autosomal Dominant Hearing Loss (DFNA1). Clin. Genet. 2017, 91, 892–901.
- Bastida, J.M.; Lozano, M.L.; Benito, R.; Janusz, K.; Palma-Barqueros, V.; Del Rey, M.; Hernández-Sánchez, J.M.; Riesco, S.; Bermejo, N.; González-García, H.; et al. Introducing High-Throughput Sequencing into Mainstream Genetic Diagnosis Practice in Inherited Platelet Disorders. Haematologica 2018, 103, 148–162.
- Ganaha, A.; Kaname, T.; Shinjou, A.; Chinen, Y.; Yanagi, K.; Higa, T.; Kondo, S.; Suzuki, M. Progressive Macrothrombocytopenia and Hearing Loss in a Large Family with DIAPH1 Related Disease. Am. J. Med. Genet. 2017, 173, 2826–2830.
- Karki, N.R.; Ajebo, G.; Savage, N.; Kutlar, A. DIAPH1 Mutation as a Novel Cause of Autosomal Dominant Macrothrombocytopenia and Hearing Loss. Acta Haematol. 2021, 144, 87–90.
- Rabbolini, D.; Connor, D.; Morel-Kopp, M.-C.; Donikian, D.; Kondo, M.; Chen, W.; Alessi, M.-C.; Stevenson, W.; Chen, V.; Joseph, J.; et al. An Integrated Approach to Inherited Platelet Disorders: Results from a Research Collaborative, the Sydney Platelet Group. Pathology 2020, 52, 243–255.
- Stritt, S.; Nurden, P.; Turro, E.; Greene, D.; Jansen, S.B.; Westbury, S.K.; Petersen, R.; Astle, W.J.; Marlin, S.; Bariana, T.K.; et al. A Gain-of-Function Variant in DIAPH1 Causes Dominant Macrothrombocytopenia and Hearing Loss. Blood 2016, 127, 2903–2914.
- Ji, H.; Lu, J.; Wang, J.; Li, H.; Lin, X. Combined Examination of Sequence and Copy Number Variations in Human Deafness Genes Improves Diagnosis for Cases of Genetic Deafness. BMC Ear Nose Throat Disord. 2014, 14, 9.
- Bione, S.; Sala, C.; Manzini, C.; Arrigo, G.; Zuffardi, O.; Banfi, S.; Borsani, G.; Jonveaux, P.; Philippe, C.; Zuccotti, M.; et al. A Human Homologue of the Drosophila Melanogaster Diaphanous Gene Is Disrupted in a Patient with Premature Ovarian Failure: Evidence for Conserved Function in Oogenesis and Implications for Human Sterility. Am. J. Hum. Genet. 1998, 62, 533–541.
- Sala, C.; Arrigo, G.; Torri, G.; Martinazzi, F.; Riva, P.; Larizza, L.; Philippe, C.; Jonveaux, P.; Sloan, F.; Labella, T.; et al. Eleven X Chromosome Breakpoints Associated with Premature Ovarian Failure (POF) Map to a 15-Mb YAC Contig Spanning Xq21. Genomics 1997, 40, 123–131.
- Marozzi, A.; Manfredini, E.; Tibiletti, M.; Furlan, D.; Villa, N.; Vegetti, W.; Crosignani, P.; Ginelli, E.; Meneveri, R.; Dalprà, L. Molecular Definition of Xq Common-Deleted Region in Patients Affected by Premature Ovarian Failure. Hum. Genet. 2000, 107, 304–311.
- Misceo, D.; Rødningen, O.K.; Barøy, T.; Sorte, H.; Mellembakken, J.R.; Strømme, P.; Fannemel, M.; Frengen, E. A Translocation between Xq21.33 and 22q13.33 Causes an Intragenic SHANK3 Deletion in a Woman with Phelan-McDermid Syndrome and Hypergonadotropic Hypogonadism. Am. J. Med. Genet. 2011, 155, 403–408.
- Genesio, R.; Mormile, A.; Licenziati, M.R.; De Brasi, D.; Leone, G.; Balzano, S.; Izzo, A.; Bonfiglio, F.; Conti, A.; Fioretti, G.; et al. Short Stature and Primary Ovarian Insufficiency Possibly Due to Chromosomal Position Effect in a Balanced X;1 Translocation. Mol. Cytogenet. 2015, 8, 50.
- Bestetti, I.; Castronovo, C.; Sironi, A.; Caslini, C.; Sala, C.; Rossetti, R.; Crippa, M.; Ferrari, I.; Pistocchi, A.; Toniolo, D.; et al. High-Resolution Array-CGH Analysis on 46, XX Patients Affected by Early Onset Primary Ovarian Insufficiency Discloses New Genes Involved in Ovarian Function. Hum. Reprod. 2019, 34, 574–583.
- Sánchez-Martínez, A.; Benito-Orejas, J.I.; Tellería-Orriols, J.J.; Alonso-Ramos, M.J. Autosomal Dominant Auditory Neuropathy and Variant DIAPH3 (c.-173C>T). Acta Otorrinolaringol. Esp. 2017, 68, 183–185.
- Schoen, C.J.; Emery, S.B.; Thorne, M.C.; Ammana, H.R.; Śliwerska, E.; Arnett, J.; Hortsch, M.; Hannan, F.; Burmeister, M.; Lesperance, M.M. Increased Activity of Diaphanous Homolog 3 (DIAPH3)/Diaphanous Causes Hearing Defects in Humans with Auditory Neuropathy and in Drosophila. Proc. Natl. Acad. Sci. USA 2010, 107, 13396–13401.
- Schneider, R.; Deutsch, K.; Hoeprich, G.J.; Marquez, J.; Hermle, T.; Braun, D.A.; Seltzsam, S.; Kitzler, T.M.; Mao, Y.; Buerger, F.; et al. DAAM2 Variants Cause Nephrotic Syndrome via Actin Dysregulation. Am. J. Hum. Genet. 2020, 107, 1113–1128.
- Law, R.; Dixon-Salazar, T.; Jerber, J.; Cai, N.; Abbasi, A.A.; Zaki, M.S.; Mittal, K.; Gabriel, S.B.; Rafiq, M.A.; Khan, V.; et al. Biallelic Truncating Mutations in FMN2, Encoding the Actin-Regulatory Protein Formin 2, Cause Nonsyndromic Autosomal-Recessive Intellectual Disability. Am. J. Hum. Genet. 2014, 95, 721–728.
- Gorukmez, O.; Gorukmez, O.; Ekici, A. A Novel Nonsense FMN2 Mutation in Nonsyndromic Autosomal Recessive Intellectual Disability Syndrome. Fetal Pediatr. Pathol. 2020, 39, 1–5.
- Marco, E.J.; Aitken, A.B.; Nair, V.P.; da Gente, G.; Gerdes, M.R.; Bologlu, L.; Thomas, S.; Sherr, E.H. Burden of de Novo Mutations and Inherited Rare Single Nucleotide Variants in Children with Sensory Processing Dysfunction. BMC Med. Genom. 2018, 11, 50.
- Perrone, M.D.; Rocca, M.S.; Bruno, I.; Faletra, F.; Pecile, V.; Gasparini, P. De Novo 911 kb Interstitial Deletion on Chromosome 1q43 in a Boy with Mental Retardation and Short Stature. Eur. J. Med. Genet. 2012, 55, 117–119.
- Almuqbil, M.; Hamdan, F.F.; Mathonnet, G.; Rosenblatt, B.; Srour, M. De Novo Deletion of FMN2 in a Girl with Mild Non-Syndromic Intellectual Disability. Eur. J. Med. Genet. 2013, 56, 686–688.
- Brown, E.J.; Schlöndorff, J.S.; Becker, D.J.; Tsukaguchi, H.; Uscinski, A.L.; Higgs, H.N.; Henderson, J.M.; Pollak, M.R. Mutations in the Formin Protein INF2 Cause Focal Segmental Glomerulosclerosis. Nat. Genet. 2010, 42, 72–76.
- Nagano, C.; Yamamura, T.; Horinouchi, T.; Aoto, Y.; Ishiko, S.; Sakakibara, N.; Shima, Y.; Nakanishi, K.; Nagase, H.; Iijima, K.; et al. Comprehensive Genetic Diagnosis of Japanese Patients with Severe Proteinuria. Sci. Rep. 2020, 10, 270.
- Varner, J.D.; Chryst-Stangl, M.; Esezobor, C.I.; Solarin, A.; Wu, G.; Lane, B.; Hall, G.; Abeyagunawardena, A.; Matory, A.; Hunley, T.E.; et al. Genetic Testing for Steroid-Resistant-Nephrotic Syndrome in an Outbred Population. Front. Pediatr. 2018, 6, 307.
- Miao, J.; Pinto, E.; Vairo, F.; Hogan, M.C.; Erickson, S.B.; El Ters, M.; Bentall, A.J.; Kukla, A.; Greene, E.L.; Hernandez, L.H.; et al. Identification of Genetic Causes of Focal Segmental Glomerulosclerosis Increases with Proper Patient Selection. Mayo Clin. Proc. 2021, 96, 2342–2353.
- Laššuthová, P.; Šafka Brožková, D.; Krůtová, M.; Neupauerová, J.; Haberlová, J.; Mazanec, R.; Dřímal, P.; Seeman, P. Improving Diagnosis of Inherited Peripheral Neuropathies through Gene Panel Analysis. Orphanet J. Rare Dis. 2016, 11, 118.
- Boyer, O.; Nevo, F.; Plaisier, E.; Funalot, B.; Gribouval, O.; Benoit, G.; Huynh Cong, E.; Arrondel, C.; Tête, M.-J.; Montjean, R.; et al. INF2 Mutations in Charcot-Marie-Tooth Disease with Glomerulopathy. N. Engl. J. Med. 2011, 365, 2377–2388.
- Roos, A.; Weis, J.; Korinthenberg, R.; Fehrenbach, H.; Häusler, M.; Züchner, S.; Mache, C.; Hubmann, H.; Auer-Grumbach, M.; Senderek, J. Inverted Formin 2-Related Charcot-Marie-Tooth Disease: Extension of the Mutational Spectrum and Pathological Findings in Schwann Cells and Axons. J. Peripher. Nerv. Syst. 2015, 20, 52–59.
- Toyota, K.; Ogino, D.; Hayashi, M.; Taki, M.; Saito, K.; Abe, A.; Hashimoto, T.; Umetsu, K.; Tsukaguchi, H.; Hayasaka, K. INF2 Mutations in Charcot-Marie-Tooth Disease Complicated with Focal Segmental Glomerulosclerosis. J. Peripher. Nerv. Syst. 2013, 18, 97–98.
- Mademan, I.; Deconinck, T.; Dinopoulos, A.; Voit, T.; Schara, U.; Devriendt, K.; Meijers, B.; Lerut, E.; De Jonghe, P.; Baets, J. De Novo INF2 Mutations Expand the Genetic Spectrum of Hereditary Neuropathy with Glomerulopathy. Neurology 2013, 81, 1953–1958.
- Barua, M.; Brown, E.J.; Charoonratana, V.T.; Genovese, G.; Sun, H.; Pollak, M.R. Mutations in the INF2 Gene Account for a Significant Proportion of Familial but Not Sporadic Focal and Segmental Glomerulosclerosis. Kidney Int. 2013, 83, 316–322.
- Snoek, R.; Nguyen, T.Q.; van der Zwaag, B.; van Zuilen, A.D.; Kruis, H.M.E.; van Gils-Verrij, L.A.; Goldschmeding, R.; Knoers, N.V.A.M.; Rookmaaker, M.B.; van Eerde, A.M. Importance of Genetic Diagnostics in Adult-Onset Focal Segmental Glomerulosclerosis. Nephron 2019, 142, 351–358.
- Riedhammer, K.M.; Braunisch, M.C.; Günthner, R.; Wagner, M.; Hemmer, C.; Strom, T.M.; Schmaderer, C.; Renders, L.; Tasic, V.; Gucev, Z.; et al. Exome Sequencing and Identification of Phenocopies in Patients with Clinically Presumed Hereditary Nephropathies. Am. J. Kidney Dis. 2020, 76, 460–470.
- Boyer, O.; Benoit, G.; Gribouval, O.; Nevo, F.; Tête, M.-J.; Dantal, J.; Gilbert-Dussardier, B.; Touchard, G.; Karras, A.; Presne, C.; et al. Mutations in INF2 Are a Major Cause of Autosomal Dominant Focal Segmental Glomerulosclerosis. J. Am. Soc. Nephrol. 2011, 22, 239–245.
- Park, E.; Lee, C.; Kim, N.K.D.; Ahn, Y.H.; Park, Y.S.; Lee, J.H.; Kim, S.H.; Cho, M.H.; Cho, H.; Yoo, K.H.; et al. Genetic Study in Korean Pediatric Patients with Steroid-Resistant Nephrotic Syndrome or Focal Segmental Glomerulosclerosis. J. Clin. Med. 2020, 9, 2013.
- Rodriguez, P.Q.; Lohkamp, B.; Celsi, G.; Mache, C.J.; Auer-Grumbach, M.; Wernerson, A.; Hamajima, N.; Tryggvason, K.; Patrakka, J. Novel INF2 Mutation p. L77P in a Family with Glomerulopathy and Charcot-Marie-Tooth Neuropathy. Pediatr. Nephrol. 2013, 28, 339–343.
- Xie, J.; Hao, X.; Azeloglu, E.U.; Ren, H.; Wang, Z.; Ma, J.; Liu, J.; Ma, X.; Wang, W.; Pan, X.; et al. Novel Mutations in the Inverted Formin 2 Gene of Chinese Families Contribute to Focal Segmental Glomerulosclerosis. Kidney Int. 2015, 88, 593–604.
- Echaniz-Laguna, A.; Latour, P. A Cryptic Splicing Mutation in the INF2 Gene Causing Charcot-Marie-Tooth Disease with Minimal Glomerular Dysfunction. J. Peripher. Nerv. Syst. 2019, 24, 120–124.
- Challis, R.C.; Ring, T.; Xu, Y.; Wong, E.K.S.; Flossmann, O.; Roberts, I.S.D.; Ahmed, S.; Wetherall, M.; Salkus, G.; Brocklebank, V.; et al. Thrombotic Microangiopathy in Inverted Formin 2-Mediated Renal Disease. J. Am. Soc. Nephrol. 2017, 28, 1084–1091.
- Bacquet, J.; Stojkovic, T.; Boyer, A.; Martini, N.; Audic, F.; Chabrol, B.; Salort-Campana, E.; Delmont, E.; Desvignes, J.-P.; Verschueren, A.; et al. Molecular Diagnosis of Inherited Peripheral Neuropathies by Targeted Next-Generation Sequencing: Molecular Spectrum Delineation. BMJ Open 2018, 8, e021632.
- Lemieux, G.; Neemeh, J.A. Charcot-Marie-Tooth Disease and Nephritis. Can. Med. Assoc. J. 1967, 97, 1193–1198.
- Yao, T.; Udwan, K.; John, R.; Rana, A.; Haghighi, A.; Xu, L.; Hack, S.; Reich, H.N.; Hladunewich, M.A.; Cattran, D.C.; et al. Integration of Genetic Testing and Pathology for the Diagnosis of Adults with FSGS. Clin. J. Am. Soc. Nephrol. 2019, 14, 213–223.
- Caridi, G.; Lugani, F.; Dagnino, M.; Gigante, M.; Iolascon, A.; Falco, M.; Graziano, C.; Benetti, E.; Dugo, M.; Del Prete, D.; et al. Novel INF2 Mutations in an Italian Cohort of Patients with Focal Segmental Glomerulosclerosis, Renal Failure and Charcot-Marie-Tooth Neuropathy. Nephrol. Dial. Transplant. 2014, 29 (Suppl. 4), iv80–iv86.
- Sadowski, C.E.; Lovric, S.; Ashraf, S.; Pabst, W.L.; Gee, H.Y.; Kohl, S.; Engelmann, S.; Vega-Warner, V.; Fang, H.; Halbritter, J.; et al. A Single-Gene Cause in 29.5% of Cases of Steroid-Resistant Nephrotic Syndrome. J. Am. Soc. Nephrol. 2015, 26, 1279–1289.
- Fu, J.; Ma, M.; Pang, M.; Yang, L.; Li, G.; Song, J.; Zhang, J. Analysis of a pedigree with autosomal dominant intermediate Charcot-Marie-Tooth disease type E and nephropathy. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2019, 36, 918–921.
- Jin, S.; Wang, W.; Wang, R.; Lv, H.; Zhang, W.; Wang, Z.; Jiao, J.; Yuan, Y. INF2 Mutations Associated with Dominant Inherited Intermediate Charcot-Marie-Tooth Neuropathy with Focal Segmental Glomerulosclerosis in Two Chinese Patients. Clin. Neuropathol. 2015, 34, 275–281.
- Park, H.J.; Kim, H.J.; Hong, Y.B.; Nam, S.H.; Chung, K.W.; Choi, B.-O. A Novel INF2 Mutation in a Korean Family with Autosomal Dominant Intermediate Charcot-Marie-Tooth Disease and Focal Segmental Glomerulosclerosis. J. Peripher. Nerv. Syst. 2014, 19, 175–179.
- Laurin, L.-P.; Lu, M.; Mottl, A.K.; Blyth, E.R.; Poulton, C.J.; Weck, K.E. Podocyte-Associated Gene Mutation Screening in a Heterogeneous Cohort of Patients with Sporadic Focal Segmental Glomerulosclerosis. Nephrol. Dial. Transplant. 2014, 29, 2062–2069.
- Wang, M.; Chun, J.; Genovese, G.; Knob, A.U.; Benjamin, A.; Wilkins, M.S.; Friedman, D.J.; Appel, G.B.; Lifton, R.P.; Mane, S.; et al. Contributions of Rare Gene Variants to Familial and Sporadic FSGS. J. Am. Soc. Nephrol. 2019, 30, 1625–1640.
- Münch, J.; Grohmann, M.; Lindner, T.H.; Bergmann, C.; Halbritter, J. Diagnosing FSGS without Kidney Biopsy—A Novel INF2-Mutation in a Family with ESRD of Unknown Origin. BMC Med. Genet. 2016, 17, 73.
- Büscher, A.K.; Celebi, N.; Hoyer, P.F.; Klein, H.-G.; Weber, S.; Hoefele, J. Mutations in INF2 May Be Associated with Renal Histology other than Focal Segmental Glomerulosclerosis. Pediatr. Nephrol. 2018, 33, 433–437.
- Braunisch, M.C.; Riedhammer, K.M.; Herr, P.-M.; Draut, S.; Günthner, R.; Wagner, M.; Weidenbusch, M.; Lungu, A.; Alhaddad, B.; Renders, L.; et al. Identification of Disease-Causing Variants by Comprehensive Genetic Testing with Exome Sequencing in Adults with Suspicion of Hereditary FSGS. Eur. J. Hum. Genet. 2021, 29, 262–270.
- Rood, I.M.; Bongers, E.M.H.F.; Lugtenberg, D.; Klein, I.H.H.T.; Steenbergen, E.J.; Wetzels, J.F.M.; Deegens, J.K.J. Familial Focal Segmental Glomerulosclerosis: Mutation in Inverted Formin 2 Mimicking Alport Syndrome. Neth. J. Med. 2016, 74, 82–85.
- Gbadegesin, R.A.; Lavin, P.J.; Hall, G.; Bartkowiak, B.; Homstad, A.; Jiang, R.; Wu, G.; Byrd, A.; Lynn, K.; Wolfish, N.; et al. Inverted Formin 2 Mutations with Variable Expression in Patients with Sporadic and Hereditary Focal and Segmental Glomerulosclerosis. Kidney Int. 2012, 81, 94–99.
- Warejko, J.K.; Tan, W.; Daga, A.; Schapiro, D.; Lawson, J.A.; Shril, S.; Lovric, S.; Ashraf, S.; Rao, J.; Hermle, T.; et al. Whole Exome Sequencing of Patients with Steroid-Resistant Nephrotic Syndrome. Clin. J. Am. Soc. Nephrol. 2018, 13, 53–62.
- Tan, W.; Lovric, S.; Ashraf, S.; Rao, J.; Schapiro, D.; Airik, M.; Shril, S.; Gee, H.Y.; Baum, M.; Daouk, G.; et al. Analysis of 24 Genes Reveals a Monogenic Cause in 11.1% of Cases with Steroid-Resistant Nephrotic Syndrome at a Single Center. Pediatr. Nephrol. 2018, 33, 305–314.
- Ogino, D.; Hashimoto, T.; Hattori, M.; Sugawara, N.; Akioka, Y.; Tamiya, G.; Makino, S.; Toyota, K.; Mitsui, T.; Hayasaka, K. Analysis of the Genes Responsible for Steroid-Resistant Nephrotic Syndrome and/or Focal Segmental Glomerulosclerosis in Japanese Patients by Whole-Exome Sequencing Analysis. J. Hum. Genet. 2016, 61, 137–141.
- Shang, S.; Peng, F.; Wang, T.; Wu, X.; Li, P.; Li, Q.; Chen, X.M. Genotype-Phenotype Correlation and Prognostic Impact in Chinese Patients with Alport Syndrome. Mol. Genet. Genom. Med. 2019, 7, e00741.
- Gribouval, O.; Boyer, O.; Hummel, A.; Dantal, J.; Martinez, F.; Sberro-Soussan, R.; Etienne, I.; Chauveau, D.; Delahousse, M.; Lionet, A.; et al. Identification of Genetic Causes for Sporadic Steroid-Resistant Nephrotic Syndrome in Adults. Kidney Int. 2018, 94, 1013–1022.
- Singh, A.; Singh, A.; Mishra, O.P.; Prasad, R.; Narayan, G.; Batra, V.V.; Tabatabaeifar, M.; Schaefer, F. Molecular Study of Childhood Steroid-Resistant Nephrotic Syndrome: A Hospital-Based Study. J. Pediatr. Genet. 2021.
- Santín, S.; Bullich, G.; Tazón-Vega, B.; García-Maset, R.; Giménez, I.; Silva, I.; Ruíz, P.; Ballarín, J.; Torra, R.; Ars, E. Clinical Utility of Genetic Testing in Children and Adults with Steroid-Resistant Nephrotic Syndrome. Clin. J. Am. Soc. Nephrol. 2011, 6, 1139–1148.
- Larsen, C.P.; Durfee, T.; Wilson, J.D.; Beggs, M.L. A Custom Targeted Next-Generation Sequencing Gene Panel for the Diagnosis of Genetic Nephropathies. Am. J. Kidney Dis. 2016, 67, 992–993.
- Zhao, W.; Ma, X.; Zhang, X.; Luo, D.; Zhang, J.; Li, M.; Ye, Z.; Peng, H. INF2 p.Arg214Cys Mutation in a Chinese Family with Rapidly Progressive Renal Failure and Follow-up of Renal Transplantation: Case Report and Literature Review. BMC Nephrol. 2021, 22, 51.
- Safarikova, M.; Stekrova, J.; Honsova, E.; Horinova, V.; Tesar, V.; Reiterova, J. Mutational Screening of Inverted Formin 2 in Adult-Onset Focal Segmental Glomerulosclerosis or Minimal Change Patients from the Czech Republic. BMC Med. Genet. 2018, 19, 147.
- Gast, C.; Pengelly, R.J.; Lyon, M.; Bunyan, D.J.; Seaby, E.G.; Graham, N.; Venkat-Raman, G.; Ennis, S. Collagen (COL4A) Mutations Are the Most Frequent Mutations Underlying Adult Focal Segmental Glomerulosclerosis. Nephrol. Dial. Transplant. 2016, 31, 961–970.
- Lipska, B.S.; Iatropoulos, P.; Maranta, R.; Caridi, G.; Ozaltin, F.; Anarat, A.; Balat, A.; Gellermann, J.; Trautmann, A.; Erdogan, O.; et al. Genetic Screening in Adolescents with Steroid-Resistant Nephrotic Syndrome. Kidney Int. 2013, 84, 206–213.
- Bullich, G.; Trujillano, D.; Santín, S.; Ossowski, S.; Mendizábal, S.; Fraga, G.; Madrid, Á.; Ariceta, G.; Ballarín, J.; Torra, R.; et al. Targeted Next-Generation Sequencing in Steroid-Resistant Nephrotic Syndrome: Mutations in Multiple Glomerular Genes May Influence Disease Severity. Eur. J. Hum. Genet. 2015, 23, 1192–1199.
- Lee, H.K.; Han, K.H.; Jung, Y.H.; Kang, H.G.; Moon, K.C.; Ha, I.S.; Choi, Y.; Cheong, H.I. Variable Renal Phenotype in a Family with an INF2 Mutation. Pediatr. Nephrol. 2011, 26, 73–76.
- Sanchez-Ares, M.; Garcia-Vidal, M.; Antucho, E.-E.; Julio, P.; Eduardo, V.-M.; Lens, X.M.; Garcia-Gonzalez, M.A. A Novel Mutation, Outside of the Candidate Region for Diagnosis, in the Inverted Formin 2 Gene Can Cause Focal Segmental Glomerulosclerosis. Kidney Int. 2013, 83, 153–159.
- Büscher, A.K.; Beck, B.B.; Melk, A.; Hoefele, J.; Kranz, B.; Bamborschke, D.; Baig, S.; Lange-Sperandio, B.; Jungraithmayr, T.; Weber, L.T.; et al. Rapid Response to Cyclosporin A and Favorable Renal Outcome in Nongenetic Versus Genetic Steroid-Resistant Nephrotic Syndrome. Clin. J. Am. Soc. Nephrol. 2016, 11, 245–253.
- Weber, S.; Büscher, A.K.; Hagmann, H.; Liebau, M.C.; Heberle, C.; Ludwig, M.; Rath, S.; Alberer, M.; Beissert, A.; Zenker, M.; et al. Dealing with the Incidental Finding of Secondary Variants by the Example of SRNS Patients Undergoing Targeted Next-Generation Sequencing. Pediatr. Nephrol. 2016, 31, 73–81.
- Dohrn, M.F.; Glöckle, N.; Mulahasanovic, L.; Heller, C.; Mohr, J.; Bauer, C.; Riesch, E.; Becker, A.; Battke, F.; Hörtnagel, K.; et al. Frequent Genes in Rare Diseases: Panel-Based next Generation Sequencing to Disclose Causal Mutations in Hereditary Neuropathies. J. Neurochem. 2017, 143, 507–522.
- Sinha, R.; Maiti, R.; Das, D.; Mandal, K. Steroid Resistant Nephrotic Syndrome with Clumsy Gait Associated with INF2 Mutation. Indian Pediatr. 2020, 57, 764.
- Wu, G.; Ruan, J.; Liu, J.; Zhang, C.; Kang, L.; Wang, J.; Zou, Y.; Song, L. Variant Spectrum of Formin Homology 2 Domain-Containing 3 Gene in Chinese Patients with Hypertrophic Cardiomyopathy. J. Am. Heart Assoc. 2021, 10, e018236.
- Huang, S.; Pu, T.; Wei, W.; Xu, R.; Wu, Y. Exome Sequencing Identifies a FHOD3 p.S527del Mutation in a Chinese Family with Hypertrophic Cardiomyopathy. J. Gene Med. 2020, 22, e3146.
- Ochoa, J.P.; Sabater-Molina, M.; García-Pinilla, J.M.; Mogensen, J.; Restrepo-Córdoba, A.; Palomino-Doza, J.; Villacorta, E.; Martinez-Moreno, M.; Ramos-Maqueda, J.; Zorio, E.; et al. Formin Homology 2 Domain Containing 3 (FHOD3) Is a Genetic Basis for Hypertrophic Cardiomyopathy. J. Am. Coll. Cardiol. 2018, 72, 2457–2467.
- Semsarian, C.; Ingles, J.; Bagnall, R.D. Revisiting Genome Sequencing Data in Light of Novel Disease Gene Associations. J. Am. Coll. Cardiol. 2019, 73, 1365–1366.
- Ochoa, J.P.; Lopes, L.R.; Perez-Barbeito, M.; Cazón-Varela, L.; de la Torre-Carpente, M.M.; Sonicheva-Paterson, N.; Uña-Iglesias, D.D.; Quinn, E.; Kuzmina-Krutetskaya, S.; Garrote, J.A.; et al. Deletions of Specific Exons of FHOD3 Detected by Next-Generation Sequencing Are Associated with Hypertrophic Cardiomyopathy. Clin. Genet. 2020, 98, 86–90.
- Hayashi, T.; Tanimoto, K.; Hirayama-Yamada, K.; Tsuda, E.; Ayusawa, M.; Nunoda, S.; Hosaki, A.; Kimura, A. Genetic Background of Japanese Patients with Pediatric Hypertrophic and Restrictive Cardiomyopathy. J. Hum. Genet. 2018, 63, 989–996.
- DeWard, A.D.; Eisenmann, K.M.; Matheson, S.F.; Alberts, A.S. The Role of Formins in Human Disease. Biochim. Biophys. Acta BBA Mol. Cell Res. 2010, 1803, 226–233.
- Randall, T.S.; Ehler, E. A Formin-g Role during Development and Disease. Eur. J. Cell Biol. 2014, 93, 205–211.
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410.
- Scott, R.P.; Quaggin, S.E. The Cell Biology of Renal Filtration. J. Cell Biol. 2015, 209, 199–210.
- Rosenberg, A.Z.; Kopp, J.B. Focal Segmental Glomerulosclerosis. Clin. J. Am. Soc. Nephrol. 2017, 12, 502–517.
- Labat-de-Hoz, L.; Alonso, M.A. The Formin INF2 in Disease: Progress from 10 Years of Research. Cell. Mol. Life Sci. 2020, 77, 4581–4600.
- Bose, B.; Cattran, D. Glomerular Diseases: FSGS. Clin. J. Am. Soc. Nephrol. 2014, 9, 626–632.
- Fogo, A.B. Causes and Pathogenesis of Focal Segmental Glomerulosclerosis. Nat. Rev. Nephrol. 2015, 11, 76–87.
- Rossor, A.M.; Polke, J.M.; Houlden, H.; Reilly, M.M. Clinical Implications of Genetic Advances in Charcot-Marie-Tooth Disease. Nat. Rev. Neurol. 2013, 9, 562–571.
- Bayraktar, S.; Nehrig, J.; Menis, E.; Karli, K.; Janning, A.; Struk, T.; Halbritter, J.; Michgehl, U.; Krahn, M.P.; Schuberth, C.E.; et al. A Deregulated Stress Response Underlies Distinct INF2-Associated Disease Profiles. J. Am. Soc. Nephrol. 2020, 31, 1296–1313.
- Mu, A.; Fung, T.S.; Kettenbach, A.N.; Chakrabarti, R.; Higgs, H.N. A Complex Containing Lysine-Acetylated Actin Inhibits the Formin INF2. Nat. Cell Biol. 2019, 21, 592–602.
- Subramanian, B.; Sun, H.; Yan, P.; Charoonratana, V.T.; Higgs, H.N.; Wang, F.; Lai, K.-M.V.; Valenzuela, D.M.; Brown, E.J.; Schlöndorff, J.S.; et al. Mice with Mutant Inf2 Show Impaired Podocyte and Slit Diaphragm Integrity in Response to Protamine-Induced Kidney Injury. Kidney Int. 2016, 90, 363–372.
- Mathis, S.; Funalot, B.; Boyer, O.; Lacroix, C.; Marcorelles, P.; Magy, L.; Richard, L.; Antignac, C.; Vallat, J.-M. Neuropathologic Characterization of INF2-Related Charcot-Marie-Tooth Disease: Evidence for a Schwann Cell Actinopathy. J. Neuropathol. Exp. Neurol. 2014, 73, 223–233.
- Tricaud, N. Myelinating Schwann Cell Polarity and Mechanically-Driven Myelin Sheath Elongation. Front. Cell. Neurosci. 2018, 11, 414.
- Takemon, Y.; Wright, V.; Davenport, B.; Gatti, D.M.; Sheehan, S.M.; Letson, K.; Savage, H.S.; Lennon, R.; Korstanje, R. Uncovering Modifier Genes of X-Linked Alport Syndrome Using a Novel Multiparent Mouse Model. J. Am. Soc. Nephrol. 2021, 32, 1961–1973.
- Schwander, M.; Kachar, B.; Müller, U. Review Series: The Cell Biology of Hearing. J. Cell Biol. 2010, 190, 9–20.
- Drummond, M.C.; Belyantseva, I.A.; Friderici, K.H.; Friedman, T.B. Actin in Hair Cells and Hearing Loss. Hear Res. 2012, 288, 89–99.
- Wallar, B.J.; Stropich, B.N.; Schoenherr, J.A.; Holman, H.A.; Kitchen, S.M.; Alberts, A.S. The Basic Region of the Diaphanous-Autoregulatory Domain (DAD) Is Required for Autoregulatory Interactions with the Diaphanous-Related Formin Inhibitory Domain. J. Biol. Chem. 2006, 281, 4300–4307.
- Miyoshi, T.; Belyantseva, I.A.; Kitajiri, S.-I.; Miyajima, H.; Nishio, S.-Y.; Usami, S.-I.; Kim, B.J.; Choi, B.Y.; Omori, K.; Shroff, H.; et al. Human Deafness-Associated Variants Alter the Dynamics of Key Molecules in Hair Cell Stereocilia F-Actin Cores. Hum. Genet. 2021.
- Lakha, R.; Montero, A.M.; Jabeen, T.; Costeas, C.C.; Ma, J.; Vizcarra, C.L. Variable Autoinhibition among Deafness-Associated Variants of Diaphanous 1 (DIAPH1). Biochemistry 2021, 60, 2320–2329.
- Ninoyu, Y.; Sakaguchi, H.; Lin, C.; Suzuki, T.; Hirano, S.; Hisa, Y.; Saito, N.; Ueyama, T. The Integrity of Cochlear Hair Cells Is Established and Maintained through the Localization of Dia1 at Apical Junctional Complexes and Stereocilia. Cell Death Dis. 2020, 11, 536.
- Schoen, C.J.; Burmeister, M.; Lesperance, M.M. Diaphanous Homolog 3 (Diap3) Overexpression Causes Progressive Hearing Loss and Inner Hair Cell Defects in a Transgenic Mouse Model of Human Deafness. PLoS ONE 2013, 8, e56520.
- Surel, C.; Guillet, M.; Lenoir, M.; Bourien, J.; Sendin, G.; Joly, W.; Delprat, B.; Lesperance, M.M.; Puel, J.-L.; Nouvian, R. Remodeling of the Inner Hair Cell Microtubule Meshwork in a Mouse Model of Auditory Neuropathy AUNA1. eNeuro 2016, 3, 0295-16.2016.
- Bae, S.-H.; Baek, J.-I.; Lee, J.D.; Song, M.H.; Kwon, T.-J.; Oh, S.-K.; Jeong, J.Y.; Choi, J.Y.; Lee, K.-Y.; Kim, U.-K. Genetic Analysis of Auditory Neuropathy Spectrum Disorder in the Korean Population. Gene 2013, 522, 65–69.
- Almazni, I.; Stapley, R.; Morgan, N.V. Inherited Thrombocytopenia: Update on Genes and Genetic Variants which May Be Associated with Bleeding. Front. Cardiovasc. Med. 2019, 6, 80.
- Becker, I.C.; Scheller, I.; Wackerbarth, L.M.; Beck, S.; Heib, T.; Aurbach, K.; Manukjan, G.; Gross, C.; Spindler, M.; Nagy, Z.; et al. Actin/Microtubule Crosstalk during Platelet Biogenesis in Mice Is Critically Regulated by Twinfilin1 and Cofilin1. Blood Adv. 2020, 4, 2124–2134.
- Italiano, J.E.; Lecine, P.; Shivdasani, R.A.; Hartwig, J.H. Blood Platelets Are Assembled Principally at the Ends of Proplatelet Processes Produced by Differentiated Megakaryocytes. J. Cell Biol. 1999, 147, 1299–1312.
- Patel, S.R. The Biogenesis of Platelets from Megakaryocyte Proplatelets. J. Clin. Investig. 2005, 115, 3348–3354.
- Potts, K.S.; Farley, A.; Dawson, C.A.; Rimes, J.; Biben, C.; de Graaf, C.; Potts, M.A.; Stonehouse, O.J.; Carmagnac, A.; Gangatirkar, P.; et al. Membrane Budding Is a Major Mechanism of in Vivo Platelet Biogenesis. J. Exp. Med. 2020, 217, e20191206.
- Pan, J.; Lordier, L.; Meyran, D.; Rameau, P.; Lecluse, Y.; Kitchen-Goosen, S.; Badirou, I.; Mokrani, H.; Narumiya, S.; Alberts, A.S.; et al. The Formin DIAPH1 (MDia1) Regulates Megakaryocyte Proplatelet Formation by Remodeling the Actin and Microtubule Cytoskeletons. Blood 2014, 124, 3967–3977.
- Zuidscherwoude, M.; Green, H.L.H.; Thomas, S.G. Formin Proteins in Megakaryocytes and Platelets: Regulation of Actin and Microtubule Dynamics. Platelets 2019, 30, 23–30.
- Ishizaki, T.; Morishima, Y.; Okamoto, M.; Furuyashiki, T.; Kato, T.; Narumiya, S. Coordination of Microtubules and the Actin Cytoskeleton by the Rho Effector MDia1. Nat. Cell Biol. 2001, 3, 8–14.
- Palazzo, A.F.; Cook, T.A.; Alberts, A.S.; Gundersen, G.G. mDia Mediates Rho-Regulated Formation and Orientation of Stable Microtubules. Nat. Cell Biol. 2001, 3, 723–729.
- Heremans, J.; Garcia-Perez, J.E.; Turro, E.; Schlenner, S.M.; Casteels, I.; Collin, R.; de Zegher, F.; Greene, D.; Humblet-Baron, S.; Lesage, S.; et al. Abnormal Differentiation of B Cells and Megakaryocytes in Patients with Roifman Syndrome. J. Allergy Clin. Immunol. 2018, 142, 630–646.
- Merico, D.; Roifman, M.; Braunschweig, U.; Yuen, R.K.C.; Alexandrova, R.; Bates, A.; Reid, B.; Nalpathamkalam, T.; Wang, Z.; Thiruvahindrapuram, B.; et al. Compound Heterozygous Mutations in the Noncoding RNU4ATAC Cause Roifman Syndrome by Disrupting Minor Intron Splicing. Nat. Commun. 2015, 6, 8718.
- Dutta, P.; Maiti, S. Expression of Multiple Formins in Adult Tissues and during Developmental Stages of Mouse Brain. Gene Expr. Patterns 2015, 19, 52–59.
- Kawabata Galbraith, K.; Kengaku, M. Multiple Roles of the Actin and Microtubule-Regulating Formins in the Developing Brain. Neurosci. Res. 2019, 138, 59–69.
- Damiani, D.; Goffinet, A.M.; Alberts, A.; Tissir, F. Lack of Diaph3 Relaxes the Spindle Checkpoint Causing the Loss of Neural Progenitors. Nat. Commun. 2016, 7, 13509.
- Thumkeo, D.; Shinohara, R.; Watanabe, K.; Takebayashi, H.; Toyoda, Y.; Tohyama, K.; Ishizaki, T.; Furuyashiki, T.; Narumiya, S. Deficiency of mDia, an Actin Nucleator, Disrupts Integrity of Neuroepithelium and Causes Periventricular Dysplasia. PLoS ONE 2011, 6, e25465.
- Eisenmann, K.M.; West, R.A.; Hildebrand, D.; Kitchen, S.M.; Peng, J.; Sigler, R.; Zhang, J.; Siminovitch, K.A.; Alberts, A.S. T Cell Responses in Mammalian Diaphanous-Related Formin MDia1 Knock-out Mice. J. Biol. Chem. 2007, 282, 25152–25158.
- Sakata, D.; Taniguchi, H.; Yasuda, S.; Adachi-Morishima, A.; Hamazaki, Y.; Nakayama, R.; Miki, T.; Minato, N.; Narumiya, S. Impaired T Lymphocyte Trafficking in Mice Deficient in an Actin-Nucleating Protein, MDia1. J. Exp. Med. 2007, 204, 2031–2038.
- Kundu, T.; Dutta, P.; Nagar, D.; Maiti, S.; Ghose, A. Coupling of Dynamic Microtubules to F-Actin by Fmn2 Regulates Chemotaxis of Neuronal Growth Cones. J. Cell Sci. 2021, 134, jcs252916.
- Sahasrabudhe, A.; Ghate, K.; Mutalik, S.; Jacob, A.; Ghose, A. Formin 2 Regulates the Stabilization of Filopodial Tip Adhesions in Growth Cones and Affects Neuronal Outgrowth and Pathfinding in Vivo. Development 2016, 143, 449–460.
- Leader, B.; Lim, H.; Carabatsos, M.J.; Harrington, A.; Ecsedy, J.; Pellman, D.; Maas, R.; Leder, P. Formin-2, Polyploidy, Hypofertility and Positioning of the Meiotic Spindle in Mouse Oocytes. Nat. Cell Biol. 2002, 4, 921–928.
- Lian, G.; Dettenhofer, M.; Lu, J.; Downing, M.; Chenn, A.; Wong, T.; Sheen, V. Filamin A- and Formin 2-Dependent Endocytosis Regulates Proliferation via the Canonical Wnt Pathway. Development 2016, 143, 4509–4520.
- Lybaek, H.; Ørstavik, K.H.; Prescott, T.; Hovland, R.; Breilid, H.; Stansberg, C.; Steen, V.M.; Houge, G. An 8.9 Mb 19p13 Duplication Associated with Precocious Puberty and a Sporadic 3.9 Mb 2q23.3q24.1 Deletion Containing NR4A2 in Mentally Retarded Members of a Family with an Intrachromosomal 19p-into-19q between-Arm Insertion. Eur. J. Hum. Genet. 2009, 17, 904–910.
- Castrillon, D.H.; Wasserman, S.A. Diaphanous Is Required for Cytokinesis in Drosophila and Shares Domains of Similarity with the Products of the Limb Deformity Gene. Development 1994, 120, 3367–3377.
- Ryley, D.A.; Wu, H.-H.; Leader, B.; Zimon, A.; Reindollar, R.H.; Gray, M.R. Characterization and Mutation Analysis of the Human Formin-2 (FMN2) Gene in Women with Unexplained Infertility. Fertil. Steril. 2005, 83, 1363–1371.
- Tšuiko, O.; Nõukas, M.; Žilina, O.; Hensen, K.; Tapanainen, J.S.; Mägi, R.; Kals, M.; Kivistik, P.A.; Haller-Kikkatalo, K.; Salumets, A.; et al. Copy Number Variation Analysis Detects Novel Candidate Genes Involved in Follicular Growth and Oocyte Maturation in a Cohort of Premature Ovarian Failure Cases. Hum. Reprod. 2016, 31, 1913–1925.
- Li, Y.; Zhang, Q.; Liu, F.; Zhang, Z.; Zou, Y.; Yang, B.; Luo, Y.; Wang, L.; Huang, O. Inhibition of Formin like 2 Promotes the Transition of Ectopic Endometrial Stromal Cells to Epithelial Cells in Adenomyosis through a MET-like Process. Gene 2019, 710, 186–192.
- Rosado, M.; Barber, C.F.; Berciu, C.; Feldman, S.; Birren, S.J.; Nicastro, D.; Goode, B.L. Critical Roles for Multiple Formins during Cardiac Myofibril Development and Repair. Mol. Biol. Cell 2014, 25, 811–827.
- Taniguchi, K.; Takeya, R.; Suetsugu, S.; Kan-o, M.; Narusawa, M.; Shiose, A.; Tominaga, R.; Sumimoto, H. Mammalian Formin Fhod3 Regulates Actin Assembly and Sarcomere Organization in Striated Muscles. J. Biol. Chem. 2009, 284, 29873–29881.
- Ehler, E. Actin-Associated Proteins and Cardiomyopathy-the “unknown” beyond Troponin and Tropomyosin. Biophys. Rev. 2018, 10, 1121–1128.
- Arimura, T.; Takeya, R.; Ishikawa, T.; Yamano, T.; Matsuo, A.; Tatsumi, T.; Nomura, T.; Sumimoto, H.; Kimura, A. Dilated Cardiomyopathy-Associated FHOD3 Variant Impairs the Ability to Induce Activation of Transcription Factor Serum Response Factor. Circ. J. 2013, 77, 2990–2996.
- Harper, A.R.; Goel, A.; Grace, C.; Thomson, K.L.; Petersen, S.E.; Xu, X.; Waring, A.; Ormondroyd, E.; Kramer, C.M.; Ho, C.Y.; et al. Common Genetic Variants and Modifiable Risk Factors Underpin Hypertrophic Cardiomyopathy Susceptibility and Expressivity. Nat. Genet. 2021, 53, 135–142.
- Esslinger, U.; Garnier, S.; Korniat, A.; Proust, C.; Kararigas, G.; Müller-Nurasyid, M.; Empana, J.-P.; Morley, M.P.; Perret, C.; Stark, K.; et al. Exome-Wide Association Study Reveals Novel Susceptibility Genes to Sporadic Dilated Cardiomyopathy. PLoS ONE 2017, 12, e0172995.
- Kan, O.M.; Takeya, R.; Abe, T.; Kitajima, N.; Nishida, M.; Tominaga, R.; Kurose, H.; Sumimoto, H. Mammalian Formin Fhod3 Plays an Essential Role in Cardiogenesis by Organizing Myofibrillogenesis. Biol. Open 2012, 1, 889–896.
- Ushijima, T.; Fujimoto, N.; Matsuyama, S.; Kan, O.M.; Kiyonari, H.; Shioi, G.; Kage, Y.; Yamasaki, S.; Takeya, R.; Sumimoto, H. The Actin-Organizing Formin Protein Fhod3 Is Required for Postnatal Development and Functional Maintenance of the Adult Heart in Mice. J. Biol. Chem. 2018, 293, 148–162.
- Heineke, J.; Molkentin, J.D. Regulation of Cardiac Hypertrophy by Intracellular Signalling Pathways. Nat. Rev. Mol. Cell Biol. 2006, 7, 589–600.
- Zhou, Q.; Wei, S.-S.; Wang, H.; Wang, Q.; Li, W.; Li, G.; Hou, J.-W.; Chen, X.-M.; Chen, J.; Xu, W.-P.; et al. Crucial Role of ROCK2-Mediated Phosphorylation and Upregulation of FHOD3 in the Pathogenesis of Angiotensin II-Induced Cardiac Hypertrophy. Hypertension 2017, 69, 1070–1083.
