Multiple myeloma is the second most common hematologic malignancy in the United States. Eventually, all myeloma patients will relapse and develop resistance to currently available agents. There is an unmet medical need to identify novel therapeutic targets. PIM kinases play an important role in myeloma pathogenesis and disease relapse.
- PIM kinase
- inhibitor
- myeloma
- resistance
- PI3K/Akt/mTOR
1. Introduction
Multiple myeloma (MM) is a hematologic malignancy characterized by the proliferation of malignant plasma cells. Precision medicine has heralded an era of change and challenge in the treatment of patients with MM. Although targeted therapies and immunotherapies have made significant advances in the personalized treatment of MM, clinicians still face the persistence of disease recurrence and drug resistance. The acquisition of anti-cancer drug resistance is a major issue with therapies in MM. Cancer cells utilize multiple intercellular and intracellular signaling cascades mediated by oncogenes such as PIM kinases to maintain cell growth and survival. In normal cells, the activity of these kinases is tightly controlled, whereas their sustained activation promotes apoptotic resistance and uncontrolled proliferation in cancer cells [1]. The complexity of the kinase molecular signaling network along with its crosstalk with alternative oncogenic signaling pathways provides ample opportunities for MM to develop productive adaptive mechanisms. PIM kinase activation has been shown to play a significant role in this bypass signaling mechanism. A better understanding of PIM kinase synergism, in addition to other signaling pathways, is important to the development of PIM inhibitors and to provide the rationale of combination therapy to improve the treatment efficacy for patients with MM.
2. Background—Expression and Regulation of PIM Kinases
3. PIM Kinase and Cancers
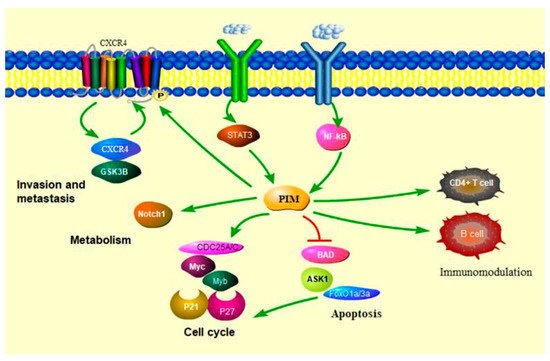
3.1. PIM Kinases in Cancer Cell Cycle
3.2. PIM Kinases in Cancer Cell Survival
3.3. PIM Kinases in Cancer Cell Metabolism
3.4. PIM Kinases and Immune Modulation
This entry is adapted from the peer-reviewed paper 10.3390/cancers13174304
References
- Warfel, N.A.; Kraft, A.S. PIM kinase (and Akt) biology and signaling in tumors. Pharmacol. Ther. 2015, 151, 41–49.
- An, N.; Lin, Y.W.; Mahajan, S.; Kellner, J.N.; Wang, Y.; Li, Z.; Kraft, A.S.; Kang, Y. Pim1 serine/threonine kinase regulates the number and functions of murine hematopoietic stem cells. Stem Cells 2013, 31, 1202–1212.
- An, N.; Kraft, A.S.; Kang, Y. Abnormal hematopoietic phenotypes in Pim kinase triple knockout mice. J. Hematol. Oncol. 2013, 6, 12.
- Le, X.; Antony, R.; Razavi, P.; Treacy, D.J.; Luo, F.; Ghandi, M.; Castel, P.; Scaltriti, M.; Baselga, J.; Garraway, L.A. Systematic Functional Characterization of Resistance to PI3K Inhibition in Breast Cancer. Cancer Discov. 2016, 6, 1134–1147.
- Iyer, R.S.; Chatham, L.; Sleigh, R.; Meek, D.W. A functional SUMO-motif in the active site of PIM1 promotes its degradation via RNF4, and stimulates protein kinase activity. Sci. Rep. 2017, 7, 3598.
- Adam, K.; Lambert, M.; Lestang, E.; Champenois, G.; Dusanter-Fourt, I.; Tamburini, J.; Bouscary, D.; Lacombe, C.; Zermati, Y.; Mayeux, P. Control of Pim2 kinase stability and expression in transformed human haematopoietic cells. Biosci. Rep. 2015, 35, e00274.
- Mologni, L.; Magistroni, V.; Casuscelli, F.; Montemartini, M.; Gambacorti-Passerini, C. The Novel PIM1 Inhibitor NMS-P645 Reverses PIM1-Dependent Effects on TMPRSS2/ERG Positive Prostate Cancer Cells And Shows Anti-Proliferative Activity in Combination with PI3K Inhibition. J. Cancer 2017, 8, 140–145.
- Hildebrand, D.; Heeg, K.; Kubatzky, K.F. Pasteurella multocida Toxin Manipulates T Cell Differentiation. Front. Microbiol. 2015, 6, 1273.
- Darici, S.; Alkhaldi, H.; Horne, G.; Jorgensen, H.G.; Marmiroli, S.; Huang, X. Targeting PI3K/Akt/mTOR in AML: Rationale and Clinical Evidence. J. Clin. Med. 2020, 9, 2934.
- Hoover, D.S.; Wingett, D.G.; Zhang, J.; Reeves, R.; Magnuson, N.S. Pim-1 protein expression is regulated by its 5′-untranslated region and translation initiation factor elF-4E. Cell Growth Differ. 1997, 8, 1371–1380.
- Keane, N.A.; Reidy, M.; Natoni, A.; Raab, M.S.; O’Dwyer, M. Targeting the Pim kinases in multiple myeloma. Blood Cancer J. 2015, 5, e325.
- Leung, C.O.; Wong, C.C.; Fan, D.N.; Kai, A.K.; Tung, E.K.; Xu, I.M.; Ng, I.O.; Lo, R.C. PIM1 regulates glycolysis and promotes tumor progression in hepatocellular carcinoma. Oncotarget 2015, 6, 10880–10892.
- Kim, O.; Jiang, T.; Xie, Y.; Guo, Z.; Chen, H.; Qiu, Y. Synergism of cytoplasmic kinases in IL6-induced ligand-independent activation of androgen receptor in prostate cancer cells. Oncogene 2004, 23, 1838–1844.
- Cervantes-Gomez, F.; Chen, L.S.; Orlowski, R.Z.; Gandhi, V. Biological effects of the Pim kinase inhibitor, SGI-1776, in multiple myeloma. Clin. Lymphoma Myeloma Leuk. 2013, 13, S317–S329.
- Koblish, H.; Li, Y.L.; Shin, N.; Hall, L.; Wang, Q.; Wang, K.; Covington, M.; Marando, C.; Bowman, K.; Boer, J.; et al. Preclinical characterization of INCB053914, a novel pan-PIM kinase inhibitor, alone and in combination with anticancer agents, in models of hematologic malignancies. PLoS ONE 2018, 13, e0199108.
- Motylewska, E.; Braun, M.; Stepien, H. High Expression of NEK2 and PIM1, but Not PIM3, Is Linked to an Aggressive Phenotype of Bronchopulmonary Neuroendocrine Neoplasms. Endocr. Pathol. 2020, 31, 264–273.
- Saurabh, K.; Scherzer, M.T.; Shah, P.P.; Mims, A.S.; Lockwood, W.W.; Kraft, A.S.; Beverly, L.J. The PIM family of oncoproteins: Small kinases with huge implications in myeloid leukemogenesis and as therapeutic targets. Oncotarget 2014, 5, 8503–8514.
- Cottage, C.T.; Bailey, B.; Fischer, K.M.; Avitabile, D.; Collins, B.; Tuck, S.; Quijada, P.; Gude, N.; Alvarez, R.; Muraski, J.; et al. Cardiac progenitor cell cycling stimulated by pim-1 kinase. Circ. Res. 2010, 106, 891–901.
- Mondello, P.; Cuzzocrea, S.; Mian, M. Pim kinases in hematological malignancies: Where are we now and where are we going? J. Hematol. Oncol. 2014, 7, 95.
- Chen, L.S.; Balakrishnan, K.; Gandhi, V. Inflammation and survival pathways: Chronic lymphocytic leukemia as a model system. Biochem. Pharmacol. 2010, 80, 1936–1945.
- Bachmann, M.; Hennemann, H.; Xing, P.X.; Hoffmann, I.; Moroy, T. The oncogenic serine/threonine kinase Pim-1 phosphorylates and inhibits the activity of Cdc25C-associated kinase 1 (C-TAK1): A novel role for Pim-1 at the G2/M cell cycle checkpoint. J. Biol. Chem. 2004, 279, 48319–48328.
- Mumenthaler, S.M.; Ng, P.Y.; Hodge, A.; Bearss, D.; Berk, G.; Kanekal, S.; Redkar, S.; Taverna, P.; Agus, D.B.; Jain, A. Pharmacologic inhibition of Pim kinases alters prostate cancer cell growth and resensitizes chemoresistant cells to taxanes. Mol. Cancer Ther. 2009, 8, 2882–2893.
- Aho, T.L.; Sandholm, J.; Peltola, K.J.; Mankonen, H.P.; Lilly, M.; Koskinen, P.J. Pim-1 kinase promotes inactivation of the pro-apoptotic Bad protein by phosphorylating it on the Ser112 gatekeeper site. FEBS Lett. 2004, 571, 43–49.
- Yan, B.; Zemskova, M.; Holder, S.; Chin, V.; Kraft, A.; Koskinen, P.J.; Lilly, M. The PIM-2 kinase phosphorylates BAD on serine 112 and reverses BAD-induced cell death. J. Biol. Chem. 2003, 278, 45358–45367.
- Gu, J.J.; Wang, Z.; Reeves, R.; Magnuson, N.S. PIM1 phosphorylates and negatively regulates ASK1-mediated apoptosis. Oncogene 2009, 28, 4261–4271.
- Hogan, C.; Hutchison, C.; Marcar, L.; Milne, D.; Saville, M.; Goodlad, J.; Kernohan, N.; Meek, D. Elevated levels of oncogenic protein kinase Pim-1 induce the p53 pathway in cultured cells and correlate with increased Mdm2 in mantle cell lymphoma. J. Biol. Chem. 2008, 283, 18012–18023.
- Santio, N.M.; Landor, S.K.; Vahtera, L.; Yla-Pelto, J.; Paloniemi, E.; Imanishi, S.Y.; Corthals, G.; Varjosalo, M.; Manoharan, G.B.; Uri, A.; et al. Phosphorylation of Notch1 by Pim kinases promotes oncogenic signaling in breast and prostate cancer cells. Oncotarget 2016, 7, 43220–43238.
- Carafa, V.; Altucci, L.; Nebbioso, A. Dual Tumor Suppressor and Tumor Promoter Action of Sirtuins in Determining Malignant Phenotype. Front. Pharmacol. 2019, 10, 38.
- Guo, S.; Mao, X.; Chen, J.; Huang, B.; Jin, C.; Xu, Z.; Qiu, S. Overexpression of Pim-1 in bladder cancer. J. Exp. Clin. Cancer Res. 2010, 29, 161.
- Xue, C.; He, Y.; Hu, Q.; Yu, Y.; Chen, X.; Chen, J.; Ren, F.; Li, J.; Ren, Z.; Cui, G.; et al. Downregulation of PIM1 regulates glycolysis and suppresses tumor progression in gallbladder cancer. Cancer Manag. Res. 2018, 10, 5101–5112.
- Daenthanasanmak, A.; Wu, Y.; Iamsawat, S.; Nguyen, H.D.; Bastian, D.; Zhang, M.; Sofi, M.H.; Chatterjee, S.; Hill, E.G.; Mehrotra, S.; et al. PIM-2 protein kinase negatively regulates T cell responses in transplantation and tumor immunity. J. Clin. Investig. 2018, 128, 2787–2801.
- Knudson, K.M.; Pritzl, C.J.; Saxena, V.; Altman, A.; Daniels, M.A.; Teixeiro, E. NFkappaB-Pim-1-Eomesodermin axis is critical for maintaining CD8 T-cell memory quality. Proc. Natl. Acad. Sci. USA 2017, 114, E1659–E1667.
- Chatterjee, S.; Chakraborty, P.; Daenthanasanmak, A.; Iamsawat, S.; Andrejeva, G.; Luevano, L.A.; Wolf, M.; Baliga, U.; Krieg, C.; Beeson, C.C.; et al. Targeting PIM Kinase with PD1 Inhibition Improves Immunotherapeutic Antitumor T-cell Response. Clin. Cancer Res. 2019, 25, 1036–1049.
- Jackson, L.J.; Pheneger, J.A.; Pheneger, T.J.; Davis, G.; Wright, A.D.; Robinson, J.E.; Allen, S.; Munson, M.C.; Carter, L.L. The role of PIM kinases in human and mouse CD4+ T cell activation and inflammatory bowel disease. Cell Immunol. 2012, 272, 200–213.
- Aho, T.L.; Lund, R.J.; Ylikoski, E.K.; Matikainen, S.; Lahesmaa, R.; Koskinen, P.J. Expression of human pim family genes is selectively up-regulated by cytokines promoting T helper type 1, but not T helper type 2, cell differentiation. Immunology 2005, 116, 82–88.
- Du, W.; Chen, T.; Ni, Y.; Hou, X.; Yu, Y.; Zhou, Q.; Wu, F.; Tang, W.; Shi, G. Role of PIM2 in allergic asthma. Mol. Med. Rep. 2017, 16, 7504–7512.
- Szydlowski, M.; Debek, S.; Prochorec-Sobieszek, M.; Szolkowska, M.; Tomirotti, A.M.; Juszczynski, P.; Szumera-Cieckiewicz, A. PIM Kinases Promote Survival and Immune Escape in Primary Mediastinal Large B-Cell Lymphoma through Modulation of JAK-STAT and NF-kappaB Activity. Am. J. Pathol. 2021, 191, 567–574.
