Most mitochondrial proteins are synthesized in the cytosol and targeted to the mitochondrial surface in a post-translational manner. The surface of the endoplasmic reticulum (ER) plays an active role in this targeting reaction. ER-associated chaperones interact with certain mitochondrial membrane protein precursors and transfer them onto receptor proteins of the mitochondrial surface in a process termed ER-SURF.
- chaperones
- contact sites
- endoplasmic reticulum
- ER-SURF
- membrane extraction
- mitochondria
- protein targeting
1. Introduction
It is the hallmark of eukaryotic cells that intracellular membranes define multiple functionally different compartments. As a consequence, many, in some cell types even most, proteins that are initially synthesized in the cytosol have to leave the cytosol to reach another cellular compartment [1][2][3]. Thus, eukaryotic cells face the challenge to specifically direct thousands of proteins to their respective position and, equally important, to remove those proteins that get stranded at foreign and inappropriate locations. While localization signals, targeting factors, receptors, and translocases for many of the residents of the different organelles were identified, we are only starting to unravel how chaperones, proteases, retro-translocases, extractors, and other “correction factors” marshal and proofread the sorting of proteins to ensure well-defined proteomes and, hence, functional cellular compartments. As if this disorder was not complicated enough, recent studies suggest that the surfaces of different organelles actively cooperate in the sorting, the targeting and the clean-up of translocation intermediates on the passage to their final residence. In particular, the relevance of the endoplasmic reticulum (ER) as the professional cellular sorting station is not restricted to proteins that enter the secretory pathway, but also supports nascent proteins destined to mitochondria, peroxisomes, lipid droplets, and chloroplasts [4][5][6][7][8][9][10]. In this review, we will provide an overview of the role of the ER in targeting and degradation of mitochondrial precursor proteins.
2. Targeting and Translocation of Mitochondrial Proteins
Mitochondria contain a small genome coding for a handful of proteins, most of which represent hydrophobic core subunits of the respiratory chain; these proteins are presumably difficult to import from the cytosol, and their expression in mitochondria allows organelle-controlled synthesis [11][12][13]. The vast majority of mitochondrial proteins, many hundreds to thousands, are encoded in the nucleus, synthesized in the cytosol and subsequently targeted and imported into the organelle (for review see [14]).
Most of these proteins are synthesized with N-terminal presequences that serve as a matrix targeting signal (MTS) [15][16]. MTSs are amphipathic helices with one positively charged and one hydrophobic surface [17]. They are recognized at the mitochondrial outer membrane by three receptor proteins, Tom20, Tom22, and Tom70 that are part of the TOM (translocase of the outer membrane) complex. However, the association of Tom70 with the TOM complex is presumably dynamic and short-lived, potentially to gather the substrates for the import pore [18][19][20]. The three receptors, which differ in their exact substrate preference, pass precursor proteins in a cooperative manner to the protein-conducting channel formed by the β-barrel protein Tom40. Tom40 serves as a general entry gate for all proteins destined to the matrix, the inner membrane, the intermembrane space (IMS), and for many outer membrane proteins. After translocation through Tom40, precursor proteins are sorted to their respective submitochondrial localizations ( Figure 1 ): Matrix and many inner membrane proteins are directed to the TIM23 (translocase of the inner membrane) complex which threads presequence-containing proteins through the inner membrane. The transfer into the matrix is promoted by the PAM (presequence translocase-associated motor) machinery by hydrolysis of matrix ATP [21] and is followed by the proteolytic removal of the presequence mediated by the mitochondrial processing peptidase, MPP.
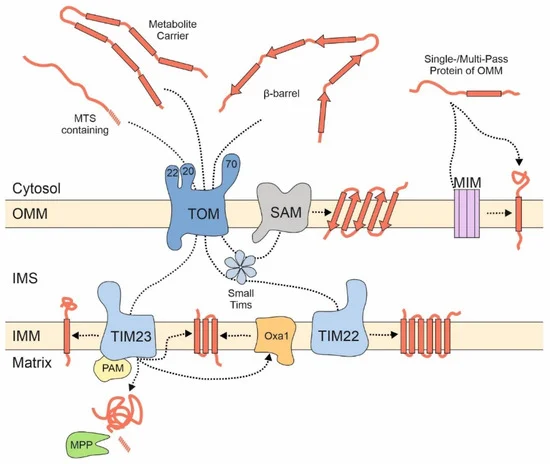 Figure 1. Different groups of mitochondrial proteins embark on different import pathways. Proteins of the matrix and many inner membrane proteins are synthesized as precursor proteins with N-terminal matrix targeting signals (MTSs) and imported via the TOM and TIM23 complexes. The PAM complex serves as motor for their translocation reaction. The mitochondrial processing peptidase (MPP) removes the MTS of most of these proteins. Metabolite carriers lack presequences and are integrated into the inner membrane by the TIM22 complex. The SAM complex integrates β-barrel proteins into the outer membrane. Many outer membrane proteins with helical transmembrane domains bypass the TOM complex but can be dependent on the MIM complex. IMM, inner mitochondrial membrane; IMS, intermembrane space; OMM, outer mitochondrial membrane.
Figure 1. Different groups of mitochondrial proteins embark on different import pathways. Proteins of the matrix and many inner membrane proteins are synthesized as precursor proteins with N-terminal matrix targeting signals (MTSs) and imported via the TOM and TIM23 complexes. The PAM complex serves as motor for their translocation reaction. The mitochondrial processing peptidase (MPP) removes the MTS of most of these proteins. Metabolite carriers lack presequences and are integrated into the inner membrane by the TIM22 complex. The SAM complex integrates β-barrel proteins into the outer membrane. Many outer membrane proteins with helical transmembrane domains bypass the TOM complex but can be dependent on the MIM complex. IMM, inner mitochondrial membrane; IMS, intermembrane space; OMM, outer mitochondrial membrane.
Carrier proteins (also referred to as the SLC25A or metabolite carrier family) are highly abundant proteins of the mitochondrial inner membrane that mediate the exchange of ATP and metabolites between mitochondria and the cytosol. They usually contain six transmembrane domains and do not contain an MTS but use internal targeting signals that are scattered across their sequence [22]. These types of proteins are first recognized on the mitochondrial surface by Tom70, which also tightly cooperates with the chaperone system of the cytosol [23][24]. Soluble chaperone complexes in the IMS, formed by small TIM proteins, and the TIM22 complex then integrate carrier proteins into the inner membrane [25][26].
The individual steps of these import reactions were elucidated by use of very powerful in vitro import assays for which radiolabeled precursor proteins were mixed with isolated mitochondria. Whereas this approach is very well suited to study protein translocation across the mitochondrial membranes, it does not reveal the initial targeting process that occurs before precursors reach the mitochondrial surface receptors.
3. Productive Targeting via the ER Surface: ER-SURF
In the context of protein biogenesis, protein targeting (the passage of nascent precursors from the ribosome to the mitochondrial surface) has to be distinguished from protein translocation (which refers to the threading of precursors through mitochondrial protein translocases) [27]. Whereas mitochondrial protein translocation was studied extensively over the last two decades, mitochondrial protein targeting is by far less understood. Many recent studies documented the general importance of the cytosolic chaperone network and the ubiquitin-proteasome-system (UPS). However, we know very little about which chaperones and which ubiquitin ligases interact with which types of precursors and how their directional movement to and subsequent release from the mitochondrial membrane is mediated. Furthermore, it is unclear whether these quality control systems usher every single precursor protein along its way to the mitochondrial surface or whether they only deal with the fraction of stranded or structurally compromised precursor proteins. Thus, the early reactions of mitochondrial protein biogenesis still await to be discovered. Several recent reviews discussed these issues in depth [27][28][29]. In this article, we therefore specifically focus on the relevance of the ER surface for mitochondrial protein biogenesis.
A large fraction of all ribosomes is bound to the surface of the ER. Ribosome-bound nascent chains that expose highly hydrophobic signal sequences or transmembrane domains are recognized by the signal recognition particle (SRP) and recruited to the ER surface. Many mitochondrial proteins contain transmembrane domains, and therefore, it is not surprising that mitochondrial membrane proteins were also identified among the SRP clients [30]. For example, transmembrane domains of the inner membrane proteins Oxa1 and Psd1 were found to be recognized by the SRP. Both proteins are synthesized with presequences and use the TIM23-mediated import pathway [31][32]. However, presumably due to the lower hydrophobicity of transmembrane domains in mitochondrial membrane proteins [33][34], the SRP is able to discriminate between secretory proteins and most mitochondrial proteins. A recent study even proposed that the major mission of the SRP system is the reliable distinction of these two large groups of cellular proteins [35]. Cotranslational binders of the nascent chain, such as Ssb1/2 chaperones and the nascent chain-associated complex (NAC), fine-tune the SRP-mediated discrimination process [36][37], which might contribute to their observed relevance for mitochondrial protein biogenesis [38][39][40][41][42]. Interestingly, the factors of the guided entry of tail-anchored proteins (GET) pathway [43] also play a role in mitochondrial protein targeting. Get3, a cytosolic chaperone and targeting factor, was found to directly interact with some mitochondrial precursors, in addition to its ER-destined clients [44].
Thus, mitochondrial proteins might initially find themselves stranded at the ER due to “mis-localization” if the SRP and cellular quality control components are not effective enough in recognizing them. However, alternatively and not mutually exclusive, some mitochondrial proteins might deliberately associate with the ER surface or be synthesized by ER-bound ribosomes, in line with a considerable number of nascent mitochondrial proteins that were observed on the ER by ribosome profiling experiments [45][46]. Regardless of whether ER targeting of nascent mitochondrial proteins is an active, intentional mechanism or a cellular mistake, cells have evolved a pathway to help such precursors make it to their final destination. This stopover-mediated targeting route to mitochondria via the ER surface was termed the ER-SURF pathway ( Figure 2 ) and the ER-bound J protein Djp1 was identified as a component that increased the efficiency of this import route . The ER-SURF model proposes that the ER surface acts as an antenna that helps to funnel precursors to mitochondria. Consistent with this idea, in in vitro experiments the addition of ER fractions increases the import rate of proteins into isolated mitochondria, an observation that can hardly be reconciled with the idea that ER binding is synonymous with non-productive mis-localization.
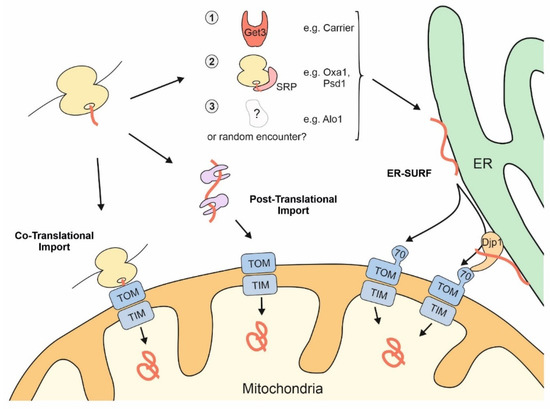 Figure 2. The surface of the ER facilitates mitochondrial targeting of proteins that use the ER-SURF pathway. Precursor proteins can reach mitochondria during, or after, their synthesis on cytosolic ribosomes. Some mitochondrial precursors are directed to the ER surface. For example, metabolite carriers were observed to be bound by Get3, a chaperone that facilitates ER-targeting of TA proteins. Some mitochondrial membrane proteins, such as Oxa1 and Psd1, are recognized by the SRP. For other mitochondrial proteins, such as Alo1, targeting factors were not identified so far. Djp1 is an ER-associated protein that facilitates the transfer of ER-bound proteins to mitochondria.
Figure 2. The surface of the ER facilitates mitochondrial targeting of proteins that use the ER-SURF pathway. Precursor proteins can reach mitochondria during, or after, their synthesis on cytosolic ribosomes. Some mitochondrial precursors are directed to the ER surface. For example, metabolite carriers were observed to be bound by Get3, a chaperone that facilitates ER-targeting of TA proteins. Some mitochondrial membrane proteins, such as Oxa1 and Psd1, are recognized by the SRP. For other mitochondrial proteins, such as Alo1, targeting factors were not identified so far. Djp1 is an ER-associated protein that facilitates the transfer of ER-bound proteins to mitochondria.
When mitochondrial import sites are limiting so that precursor proteins accumulate outside of mitochondria, a large number of precursors of mitochondrial membrane proteins were found to associate with the ER surface [47] and to induce the unfolded protein response pathway of the ER [48]. Since these precursor proteins, in particular those of the carriers, have a highly toxic potential [24][49][50], ER binding might serve as a safeguard mechanism [51]. This is because the ER surface is coated by a number of cellular chaperones. For example, Ydj1, the most abundant DnaJ-type co-chaperone of yeast cells, is tethered to the ER surface by a farnesyl anchor [52].
4. Contact Sites of the ER and Mitochondria
A function for the close cooperation of ER and mitochondria via membrane contact sites was identified three decades ago when MAMs were first purified [53][54]. The MAM was described as a membranous fraction that was inseparable even from highly purified mitochondria and found to play a role in phospholipid biosynthesis. However, molecular tethers that connect mitochondria to the ER remained unclear until the ERMES (ER mitochondria encounter structure) complex was identified in budding yeast in 2009 [55]. This complex consists of the four structural components Mmm1, Mdm12, Mdm34, and Mdm10, which form a chain-like bridge holding the two organelles in close proximity [55][56][57][58]. ERMES is mainly involved in the transfer of phophatidylserine from the ER to mitochondria where it is converted to phosphatidylethanolamine. Deletion of any ERMES component leads to a decrease in the rate of phosphatidylethanolamine synthesis and the overall levels of cardiolipin in mitochondria and to a collapse of the mitochondrial network [59][60][61][62][63]. Moreover, ERMES promotes the formation of mitochondria-derived compartments (MDCs) [6], defines the position of intra-mitochondrial complexes such as nucleoids, the MICOS (mitochondrial contact site and cristae organizing center), and the coenzyme Q synthome.
This entry is adapted from the peer-reviewed paper 10.3390/ijms22179655
References
- Weill, U.; Yofe, I.; Sass, E.; Stynen, B.; Davidi, D.; Natarajan, J.; Ben-Menachem, R.; Avihou, Z.; Goldman, O.; Harpaz, N.; et al. Genome-wide SWAp-Tag yeast libraries for proteome exploration. Nat. Methods 2018, 15, 617–622.
- Foster, L.J.; de Hoog, C.L.; Zhang, Y.; Zhang, Y.; Xie, X.; Mootha, V.K.; Mann, M. A mammalian organelle map by protein correlation profiling. Cell 2006, 125, 187–199.
- Huh, W.K.; Falvo, J.V.; Gerke, L.C.; Carroll, A.S.; Howson, R.W.; Weissman, J.S.; O’Shea, E.K. Global analysis of protein localization in budding yeast. Nature 2003, 425, 686–691.
- Herker, E.; Vieyres, G.; Beller, M.; Krahmer, N.; Bohnert, M. Lipid Droplet Contact Sites in Health and Disease. Trends Cell Biol. 2021, 31, 345–358.
- Hansen, K.G.; Aviram, N.; Laborenz, J.; Bibi, C.; Meyer, M.; Spang, A.; Schuldiner, M.; Herrmann, J.M. An ER surface retrieval pathway safeguards the import of mitochondrial membrane proteins in yeast. Science 2018, 361, 1118–1122.
- English, A.M.; Schuler, M.H.; Xiao, T.; Kornmann, B.; Shaw, J.M.; Hughes, A.L. ER-mitochondria contacts promote mitochondrial-derived compartment biogenesis. J. Cell Biol. 2020, 219.
- Baune, M.C.; Lansing, H.; Fischer, K.; Meyer, T.; Charton, L.; Linka, N.; von Schaewen, A. The Arabidopsis Plastidial Glucose-6-Phosphate Transporter GPT1 is Dually Targeted to Peroxisomes via the Endoplasmic Reticulum. Plant Cell 2020, 32, 1703–1726.
- Dederer, V.; Khmelinskii, A.; Huhn, A.G.; Okreglak, V.; Knop, M.; Lemberg, M.K. Cooperation of mitochondrial and ER factors in quality control of tail-anchored proteins. Elife 2019, 8.
- Villarejo, A.; Buren, S.; Larsson, S.; Dejardin, A.; Monne, M.; Rudhe, C.; Karlsson, J.; Jansson, S.; Lerouge, P.; Rolland, N.; et al. Evidence for a protein transported through the secretory pathway en route to the higher plant chloroplast. Nat. Cell Biol. 2005, 7, 1224–1231.
- Radhamony, R.N.; Theg, S.M. Evidence for an ER to Golgi to chloroplast protein transport pathway. Trends Cell Biol. 2006, 16, 385–387.
- Bjorkholm, P.; Harish, A.; Hagstrom, E.; Ernst, A.M.; Andersson, S.G. Mitochondrial genomes are retained by selective constraints on protein targeting. Proc. Natl. Acad. Sci. USA 2015, 112, 10154–10161.
- Bertgen, L.; Muhlhaus, T.; Herrmann, J.M. Clingy genes: Why were genes for ribosomal proteins retained in many mitochondrial genomes? Biochim. Biophys. Acta-Bioenerg. 2020, 1861, 148275.
- Allen, J.F. Why chloroplasts and mitochondria retain their own genomes and genetic systems: Colocation for redox regulation of gene expression. Proc. Natl. Acad. Sci. USA 2015, 112, 10231–10238.
- Chacinska, A.; Koehler, C.M.; Milenkovic, D.; Lithgow, T.; Pfanner, N. Importing mitochondrial proteins: Machineries and mechanisms. Cell 2009, 138, 628–644.
- Vögtle, F.N.; Wortelkamp, S.; Zahedi, R.P.; Becker, D.; Leidhold, C.; Gevaert, K.; Kellermann, J.; Voos, W.; Sickmann, A.; Pfanner, N.; et al. Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell 2009, 139, 428–439.
- Garg, S.G.; Gould, S.B. The Role of Charge in Protein Targeting Evolution. Trends Cell Biol. 2016, 26, 894–905.
- von Heijne, G. Mitochondrial targeting sequences may form amphiphilic helices. EMBO J. 1986, 5, 1335–1342.
- Araiso, Y.; Tsutsumi, A.; Qiu, J.; Imai, K.; Shiota, T.; Song, J.; Lindau, C.; Wenz, L.S.; Sakaue, H.; Yunoki, K.; et al. Structure of the mitochondrial import gate reveals distinct preprotein paths. Nature 2019, 575, 395–401.
- Shiota, T.; Imai, K.; Qiu, J.; Hewitt, V.L.; Tan, K.; Shen, H.H.; Sakiyama, N.; Fukasawa, Y.; Hayat, S.; Kamiya, M.; et al. Molecular architecture of the active mitochondrial protein gate. Science 2015, 349, 1544–1548.
- Kreimendahl, S.; Rassow, J. The Mitochondrial Outer Membrane Protein Tom70-Mediator in Protein Traffic, Membrane Contact Sites and Innate Immunity. Int. J. Mol. Sci. 2020, 21, 7262.
- Mokranjac, D. How to get to the other side of the mitochondrial inner membrane—The protein import motor. Biol. Chem. 2020, 401, 723–736.
- Horten, P.; Colina-Tenorio, L.; Rampelt, H. Biogenesis of Mitochondrial Metabolite Carriers. Biomolecules 2020, 10, 1008.
- Young, J.C.; Hoogenraad, N.J.; Hartl, F.U. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell 2003, 112, 41–50.
- Backes, S.; Bykov, Y.S.; Flohr, T.; Raschle, M.; Zhou, J.; Lenhard, S.; Kramer, L.; Muhlhaus, T.; Bibi, C.; Jann, C.; et al. The chaperone-binding activity of the mitochondrial surface receptor Tom70 protects the cytosol against mitoprotein-induced stress. Cell Rep. 2021, 35, 108936.
- Sirrenberg, C.; Bauer, M.F.; Guiard, B.; Neupert, W.; Brunner, M. Import of carrier proteins into the mitochondrial inner membrane mediated by Tim22. Nature 1996, 384, 582–585.
- Koehler, C.M.; Jarosch, E.; Tokatlidis, K.; Schmid, K.; Schweyen, R.J.; Schatz, G. Import of mitochondrial carrier proteins mediated by essential proteins of the intermembrane space. Science 1998, 279, 369–373.
- Bykov, Y.S.; Rapaport, D.; Herrmann, J.M.; Schuldiner, M. Cytosolic Events in the Biogenesis of Mitochondrial Proteins. Trends Biochem. Sci. 2020, 45, 650–667.
- Song, J.; Herrmann, J.M.; Becker, T. Quality control of the mitochondrial proteome. Nat. Rev. Mol. Cell Biol. 2020.
- Avendano-Monsalve, M.C.; Ponce-Rojas, J.C.; Funes, S. From cytosol to mitochondria: The beginning of a protein journey. Biol. Chem. 2020, 401, 645–661.
- Chartron, J.W.; Hunt, K.C.; Frydman, J. Cotranslational signal-independent SRP preloading during membrane targeting. Nature 2016, 536, 224–228.
- Herrmann, J.M.; Neupert, W.; Stuart, R.A. Insertion into the mitochondrial inner membrane of a polytopic protein, the nuclear encoded Oxa1p. EMBO J. 1997, 16, 2217–2226.
- Horvath, S.E.; Bottinger, L.; Vogtle, F.N.; Wiedemann, N.; Meisinger, C.; Becker, T.; Daum, G. Processing and topology of the yeast mitochondrial phosphatidylserine decarboxylase 1. J. Biol. Chem. 2012, 287, 36744–36755.
- Pogozheva, I.D.; Lomize, A.L. Evolution and adaptation of single-pass transmembrane proteins. Biochim. Biophys. Acta Biomembr. 2018, 1860, 364–377.
- Perez-Martinez, X.; Vazquez-Acevedo, M.; Tolkunova, E.; Funes, S.; Claros, M.G.; Davidson, E.; King, M.P.; Gonzalez-Halphen, D. Unusual location of a mitochondrial gene. Subunit III of cytochrome C oxidase is encoded in the nucleus of Chlamydomonad algae. J. Biol. Chem. 2000, 275, 30144–30152.
- Costa, E.A.; Subramanian, K.; Nunnari, J.; Weissman, J.S. Defining the physiological role of SRP in protein-targeting efficiency and specificity. Science 2018, 359, 689–692.
- Doring, K.; Ahmed, N.; Riemer, T.; Suresh, H.G.; Vainshtein, Y.; Habich, M.; Riemer, J.; Mayer, M.P.; O’Brien, E.P.; Kramer, G.; et al. Profiling Ssb-Nascent Chain Interactions Reveals Principles of Hsp70-Assisted Folding. Cell 2017, 170, 298–311.e20.
- Hsieh, H.H.; Lee, J.H.; Chandrasekar, S.; Shan, S.O. A ribosome-associated chaperone enables substrate triage in a cotranslational protein targeting complex. Nat. Commun. 2020, 11, 5840.
- Ponce-Rojas, J.C.; Avendano-Monsalve, M.C.; Yanez-Falcon, A.R.; Jaimes-Miranda, F.; Garay, E.; Torres-Quiroz, F.; DeLuna, A.; Funes, S. alphabeta’-NAC cooperates with Sam37 to mediate early stages of mitochondrial protein import. FEBS J. 2017, 284, 814–830.
- Fünfschilling, U.; Rospert, S. Nascent polypeptide-associated complex stimulates protein import into yeast mitochondria. Mol. Biol. Cell 1999, 10, 3289–3299.
- Gamerdinger, M.; Hanebuth, M.A.; Frickey, T.; Deuerling, E. The principle of antagonism ensures protein targeting specificity at the endoplasmic reticulum. Science 2015, 348, 201–207.
- Dunn, C.D.; Jensen, R.E. Suppression of a defect in mitochondrial protein import identifies cytosolic proteins required for viability of yeast cells lacking mitochondrial DNA. Genetics 2003, 165, 35–45.
- Lesnik, C.; Cohen, Y.; Atir-Lande, A.; Schuldiner, M.; Arava, Y. OM14 is a mitochondrial receptor for cytosolic ribosomes that supports co-translational import into mitochondria. Nat. Commun. 2014, 5, 5711.
- Schuldiner, M.; Metz, J.; Schmid, V.; Denic, V.; Rakwalska, M.; Schmitt, H.D.; Schwappach, B.; Weissman, J.S. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell 2008, 134, 634–645.
- Vitali, D.G.; Sinzel, M.; Bulthuis, E.P.; Kolb, A.; Zabel, S.; Mehlhorn, D.G.; Figueiredo Costa, B.; Farkas, A.; Clancy, A.; Schuldiner, M.; et al. The GET pathway can increase the risk of mitochondrial outer membrane proteins to be mistargeted to the ER. J. Cell Sci. 2018, 131.
- Williams, C.C.; Jan, C.H.; Weissman, J.S. Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science 2014, 346, 748–751.
- Jan, C.H.; Williams, C.C.; Weissman, J.S. Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science 2014, 346, 1257521.
- Shakya, V.P.; Barbeau, W.A.; Xiao, T.; Knutson, C.S.; Schuler, M.H.; Hughes, A.L. A nuclear-based quality control pathway for non-imported mitochondrial proteins. Elife 2021, 10.
- Knöringer, K.; Groh, C.; Krämer, L.; Stein, K.C.; Hansen, K.G.; Herrmann, J.M.; Frydman, J.; Boos, F. The unfolded protein response of the endoplasmic reticulum supports mitochondrial biogenesis by buffering non-imported proteins. bioRxiv 2021.
- Schlagowski, A.M.; Knoringer, K.; Morlot, S.; Sanchez Vicente, A.; Flohr, T.; Kramer, L.; Boos, F.; Khalid, N.; Ahmed, S.; Schramm, J.; et al. Increased levels of mitochondrial import factor Mia40 prevent the aggregation of polyQ proteins in the cytosol. EMBO J. 2021, e107913.
- Wang, X.; Chen, X.J. A cytosolic network suppressing mitochondria-mediated proteostatic stress and cell death. Nature 2015, 524, 481–484.
- Xiao, T.; Shakya, V.P.S.; Hughes, A.L. The GET pathway safeguards against non-imported mitochondrial protein stress. bioRxiv 2020.
- Caplan, A.J.; Tsai, J.; Casey, P.J.; Douglas, M.G. Farnesylation of YDJ1p is required for function at elevated growth temperatures in Saccharomcyes cerevisiae. J. Biol. Chem. 1992, 267, 18890–18895.
- Rusinol, A.E.; Cui, Z.; Chen, M.H.; Vance, J.E. A unique mitochondria-associated membrane fraction from rat liver has a high capacity for lipid synthesis and contains pre-Golgi secretory proteins including nascent lipoproteins. J. Biol. Chem. 1994, 269, 27494–27502.
- Vance, J.E. Phospholipid synthesis in a membrane fraction associated with mitochondria. J. Biol. Chem. 1990, 265, 7248–7256.
- Kornmann, B.; Currie, E.; Collins, S.R.; Schuldiner, M.; Nunnari, J.; Weissman, J.S.; Walter, P. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science 2009, 325, 477–481.
- Stroud, D.A.; Oeljeklaus, S.; Wiese, S.; Bohnert, M.; Lewandrowski, U.; Sickmann, A.; Guiard, B.; van der Laan, M.; Warscheid, B.; Wiedemann, N. Composition and topology of the endoplasmic reticulum-mitochondria encounter structure. J. Mol. Biol. 2011, 413, 743–750.
- Kawano, S.; Tamura, Y.; Kojima, R.; Bala, S.; Asai, E.; Michel, A.H.; Kornmann, B.; Riezman, I.; Riezman, H.; Sakae, Y.; et al. Structure-function insights into direct lipid transfer between membranes by Mmm1-Mdm12 of ERMES. J. Cell Biol. 2018, 217, 959–974.
- Jeong, H.; Park, J.; Jun, Y.; Lee, C. Crystal structures of Mmm1 and Mdm12-Mmm1 reveal mechanistic insight into phospholipid trafficking at ER-mitochondria contact sites. Proc. Natl. Acad. Sci. USA 2017, 114, E9502–E9511.
- Sogo, L.F.; Yaffe, M.P. Regulation of mitochondrial morphology and inheritance by Mdm10p, a protein of the mitochondrial outer membrane. J. Cell Biol. 1994, 130, 1361–1373.
- Burgess, S.M.; Delannoy, M.; Jensen, R.E. MMM1 encodes a mitochondrial outer membrane protein essential for establishing and maintaining the structure of yeast mitochondria. J. Cell Biol. 1994, 126, 1375–1391.
- Berger, K.L.; Sogo, L.F.; Yaffe, M.P. Mdm12p, a component required for mitochondrial inheritance that is conserved between budding and fission yeast. J. Cell Biol. 1997, 136, 545–553.
- Dimmer, K.S.; Fritz, S.; Fuchs, F.; Messerschmitt, M.; Weinbach, N.; Neupert, W.; Westermann, B. Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Mol. Biol. Cell 2002, 13, 847–853.
- Murley, A.; Lackner, L.L.; Osman, C.; West, M.; Voeltz, G.K.; Walter, P.; Nunnari, J. ER-associated mitochondrial division links the distribution of mitochondria and mitochondrial DNA in yeast. Elife 2013, 2, e00422.
